Research Topics - Membrane Biology
Exchange of materials and information between a living cell and its environment is mediated and regulated by membrane proteins. These proteins are involved in the regulation of electrical activity of the cell, transport of water and water soluble materials across the membrane, and production of ATP. Membrane receptors are the sites for detection informational signals, such as neurotransmitters and hormones, light, and even mechanical stress. The atomic-resolution structures of a few membrane proteins have been solved, and recent advances in structure determination of membrane proteins promise more structure to be solved soon. Our group studies the mechanism of function of membrane proteins with various computational methodologies. The proteins are simulated in their natural environment, i.e., embedded in fully hydrated patches of lipid bilayers and under constant pressure and temperature conditions. The main objective is to understand how specific structural motifs and/or chemical interactions in a protein play a role in its function.
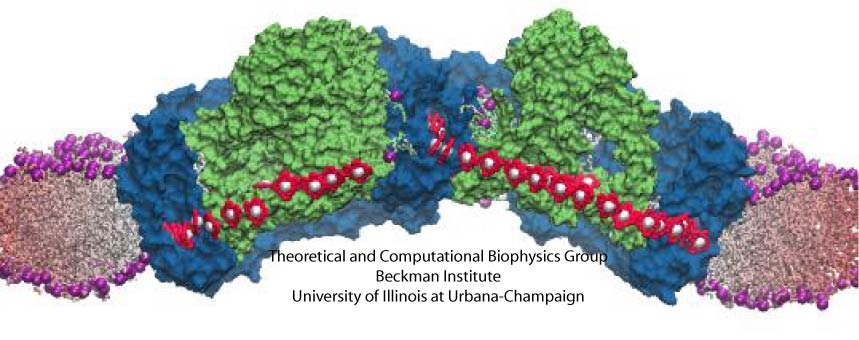
image size:
440.0KB
made with VMD

image size:
661.9KB
made with VMD
image size:
82.5KB
made with VMD
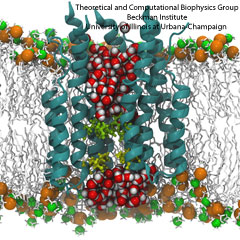
Aquaporins are membrane water channels that play critical roles in controlling the water contents of cells. These channels are widely distributed in all kingdoms of life, including bacteria, plants, and mammals. More than ten different aquaporins have been found in human body, and several diseases, such as congenital cataracts and nephrogenic diabetes insipidus, are connected to the impaired function of these channels. They form tetramers in the cell membrane, and facilitate the transport of water and, in some cases, other small solutes across the membrane. However, the water pores are completely impermeable to charged species, such as protons, a remarkable property that is critical for the conservation of membrane's electrochemical potential, but paradoxical at the same time, since protons can usually be transfered readily through water molecules. The results of our simulations have now provided new insight into the mechanism underlying this fascinating property. Water molecules passing the channel are forced, by the protein's electrostatic forces, to flip at the center of the channel (see the animation), thereby breaking the alternative donor-acceptor arrangement that is necessary for proton translocation (read the complete story in our Science paper).
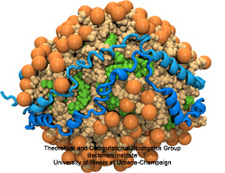
image size:
496.6KB
made with VMD
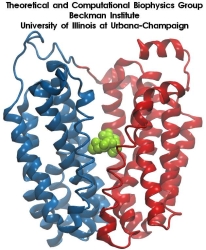
image size:
215.8KB
made with VMD
In a biological cell, membrane channels act like miniature valves regulating the flow of ions and other solutes between intracellular compartments and across the cell's boundary. Assembled in complex circuits, they generate, transmit, and amplify signals orchestrating cell function. To investigate how membrane channels work, life scientists, using an extremely fine pipette, isolate a tiny patch of a cell membrane and, in so-called patch clamp measurements, determine electric currents in response to applied electric potentials. Dramatic increase in computational power and its efficient utilization by NAMD allows one today to reproduce such studies computationally, calculating the permeability of a membrane channel to ions and water directly from its atomic structure. In what is one of the largest molecular dynamics simulation to date, described in a recent paper as well as on our web site (here), one copy of the membrane channel alpha-hemolysin, submerged in a lipid membrane and water, was subject to an external electric field that drove ions and water through the channel. The calculations produced also an image of the electrostatic potential across the channel (see figure).
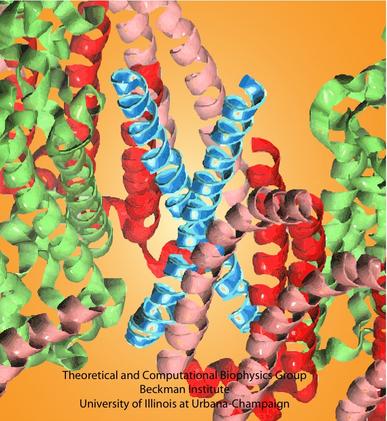
image size:
115.2KB
made with VMD
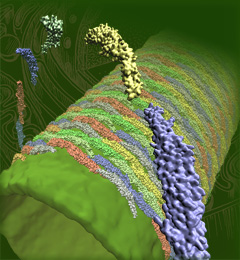
image size:
1.3MB
Olga Svinarski and VMD
The cells of higher life forms, so-called eukaryotic cells, are subdivided through many internal membranes made of lipid bilayers. The internal membranes assume numerous shapes, like spheres, tubes or parallel sheets. Outside of cells, biological membranes adopt usually flat shapes and the question arises, how do eukaryotic cells sculpt their inner membranes? The question of flat membrane sculpting is particularly interesting also as mature cells constantly produce new membrane shapes, for example spherical vesicles filled with certain biomolecules destined for release into the extracellular space, a process called exocytosis. The cell has many mechanisms available for sculpting its membranes, one of them relying on proteins called BAR domains that act from the surface of lipid bilayers. Molecular modeling with NAMD and VMD has provided valuable views of BAR domains at work in case of the so-called N-BAR family (see the earlier highlights Protein Teamwork, Jun 2009 and Proteins Sculpting Cell Interior, Sep 2008). Researchers report now an extension of the earlier studies to the F-BAR domain family of membrane sculpting proteins. The new modeling work is particularly exciting as it can be directly compared to electron microscopy images of membrane tubes sculpted from flat membranes in experiments done outside of cells. The new studies reveal how F-BAR domains sculpt tubular membranes through the shape of dimerized domains and through F-BAR domains not acting individually, but as an army of F-BAR domains adopting an ordered formation on one side of the membrane. More on our F-BAR domain web page.
All living systems contain proteins whose job is to move ions across a lipid membrane. Even viruses encode ion transport proteins, which they need to complete their lifecycle and release themselves from infected cells. Such proteins, called viroporins, usually consist of small subunits of one or two helices that can self-assemble in a lipid bilayer into a pore-like structure. Although in some cases, the resulting structures resemble the well-ordered, selective ion channels in higher organisms, often they take on a more disordered character, forming pores with variable numbers of subunits, which adapt their structure and behavior to the environment in which they find themselves. This inherent flexibility and disorder makes it very difficult to produce high-resolution crystal structures of viroporins, which is unfortunate, since they could offer attractive drug targets for new antiviral therapies. Computational modelling and molecular dynamics simulations can help fill in the gaps in our structural knowledge of viroporins, and provide plausible 3-D models for visualization and drug design. In a recent publication, scientists published models of the p7 viroporin found in Hepatitis C virus. MD simulations of these models revealed that p7 can form stable pores with 4 to 7 subunits, with a bias towards 6 or 7 subunits, and that the p7 oligomers are highly flexible in adapting to different membrane thicknesses. These simulations also suggested that specific amino acids in certain places in the structure could play a role in controlling the ion permeability of p7. More details can be found on our p7 website.
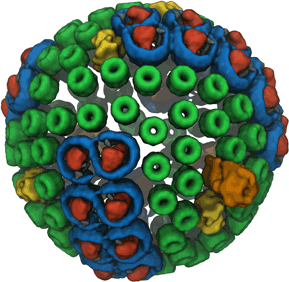
image size:
740.7KB
made with VMD
Photosynthesis, the main source of energy for all life, is performed by an intricate assembly of hundreds of proteins, which harvest, transfer, convert, and store solar energy. The simplest such light harvesting machine is the purple bacterial photosynthetic unit (PSU), which performs anoxygenic photosynthesis and is significantly simpler and evolutionarily more primitive than its counterpart found in plants. The bacterial PSU is organized in the form of a pseudo-spherical membrane domain of approximately 60 nm diameter called a chromatophore vesicle.