Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
Proteins are molecular machines inside living cells which carry out tasks from chemical synthesis to cellular motion. Each protein is a linear polymers
made of units that each are one of twenty amino acids, A(1) to A(20),
for example from a sequence A(5) - A(12) - A(1) - A(5) - ... - A(17).
Random sequences lead to disordered, non-functional proteins.
A central feature of life is that evolution has given rise to
protein sequences which fold into functional proteins. The
process through which the disordered linear polymer folds
into its final structure, however, remains mysterious, since it
is very difficult to observe experimentally. Unfortunately,
simulations of protein folding are very computationally
demanding and have uncertain outcomes (see the May
2009 highlight), which has historically
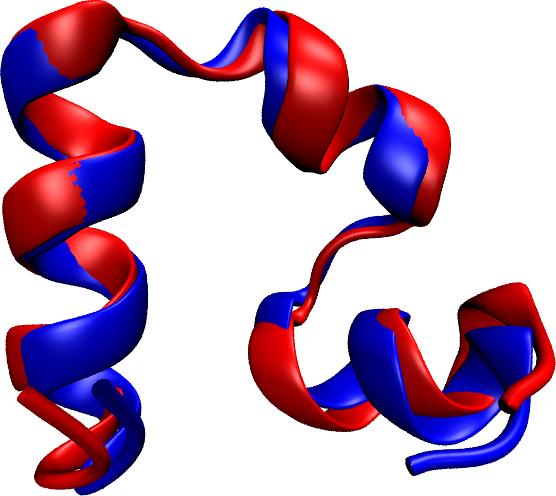
limited their utility. Now, researchers have used NAMD to simulate the
complete folding process of a small protein, villin headpiece, as reported
recently. In a series of atomic resolution molecular dynamics
simulations, covering a total of 50 microseconds, multiple
folding events were observed. Importantly, the simulations
provided a glimpse at the prevalent intermediate conformations
visited by villin during folding. A key transition between two
intermediates was recognized as rate-limiting in the villin
folding process. More information is available at our protein folding
website.