Highlights of our Work
2026 | 2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
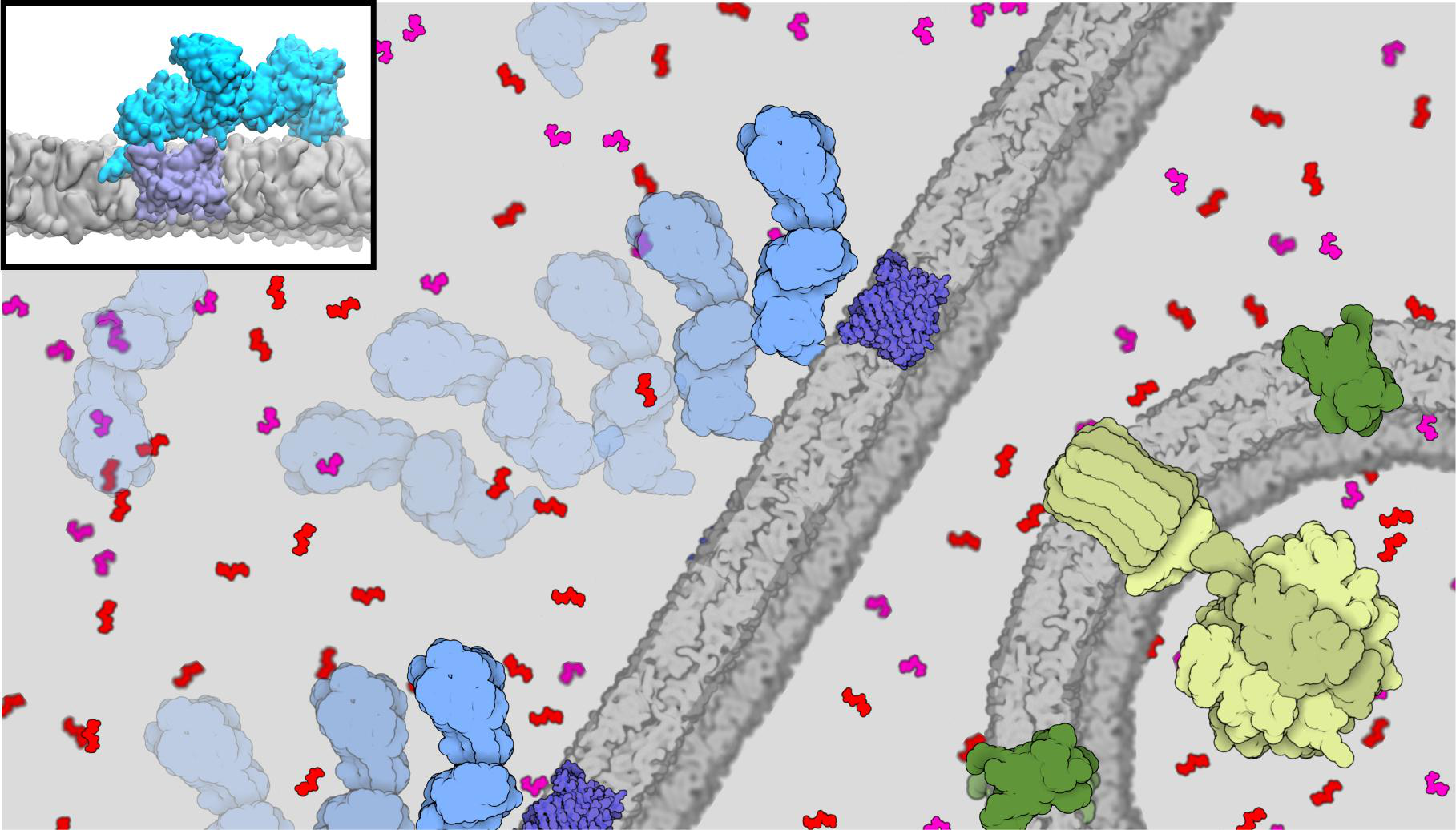
The SARS-CoV-2 replication transcription complex (RTC) is responsible for replicating and transcribing the viral mRNA inside a human cell, and a key target for pharmaceutical treatments for COVID-19. As a part of a talented multi-institutional interdisciplinary team, the center researchers conducted a multiscale study on the SARS-CoV-2 RTC, aiming at its better structural and mechanistic understanding. The study was nominated for the 2021 Gordon Bell Special Prize for COVID-19 Research, considered the "Nobel Prize for high performance computing". An innovative multi-site scientific workflow that combines the best attributes of cryo-EM imaging, all-atom molecular dynamics, and fluctuating finite element analysis with AI methods was developed to elucidate the structure and dynamics for the RTC previously inaccessible by other techniques. Team members from the Center provided key methodological advances in NAMD and VMD for the study. Read more in Argonne press release, NVIDIA press release and watch team presentation at Supercomputing 2021.
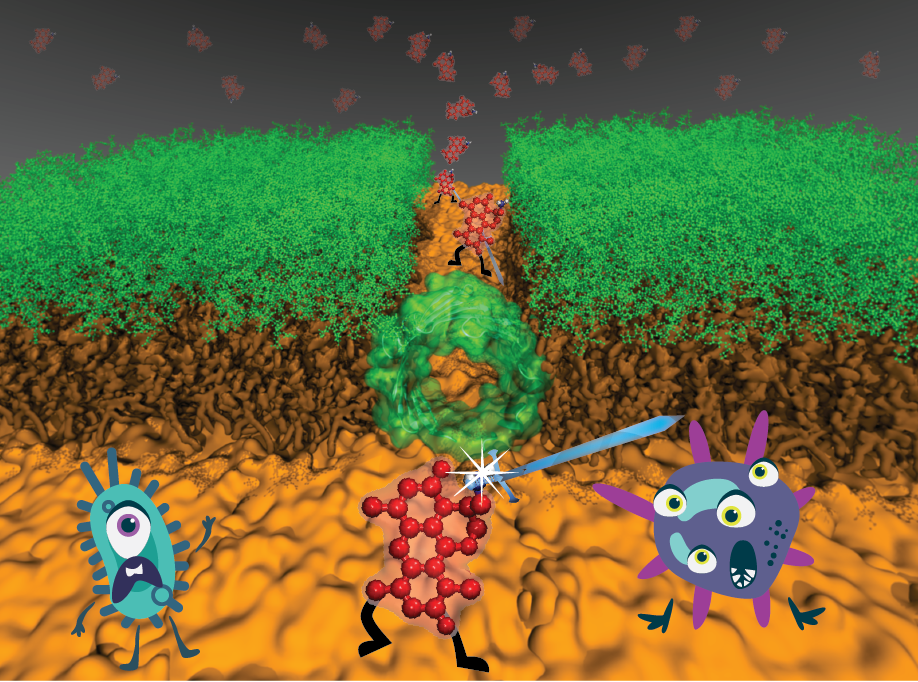
Antibiotic resistance is one of the biggest threats to global health, food security, and development today. In this context, developing antibiotics against resistant Gram-negative pathogens has proven particularly difficult due to their special dense membrane that prevents molecules from penetrating the cell. A breakthrough discovery by the Hergenrother lab in UI Chemistry showed that addition of a positive charge can help the antibiotic overcome this barrier. Using newly developed method used in NAMD simulations, the center researchers showed how the introduced charge helps antibiotics cross the membrane. Center researchers also provide mechanistic insight on gating transitions for proteins responsible for antibiotic permeation. The obtained microscopic picture could help design effective antibiotics against resistant bacteria. Read more in Chemical Science, PNAS and in UI press release.
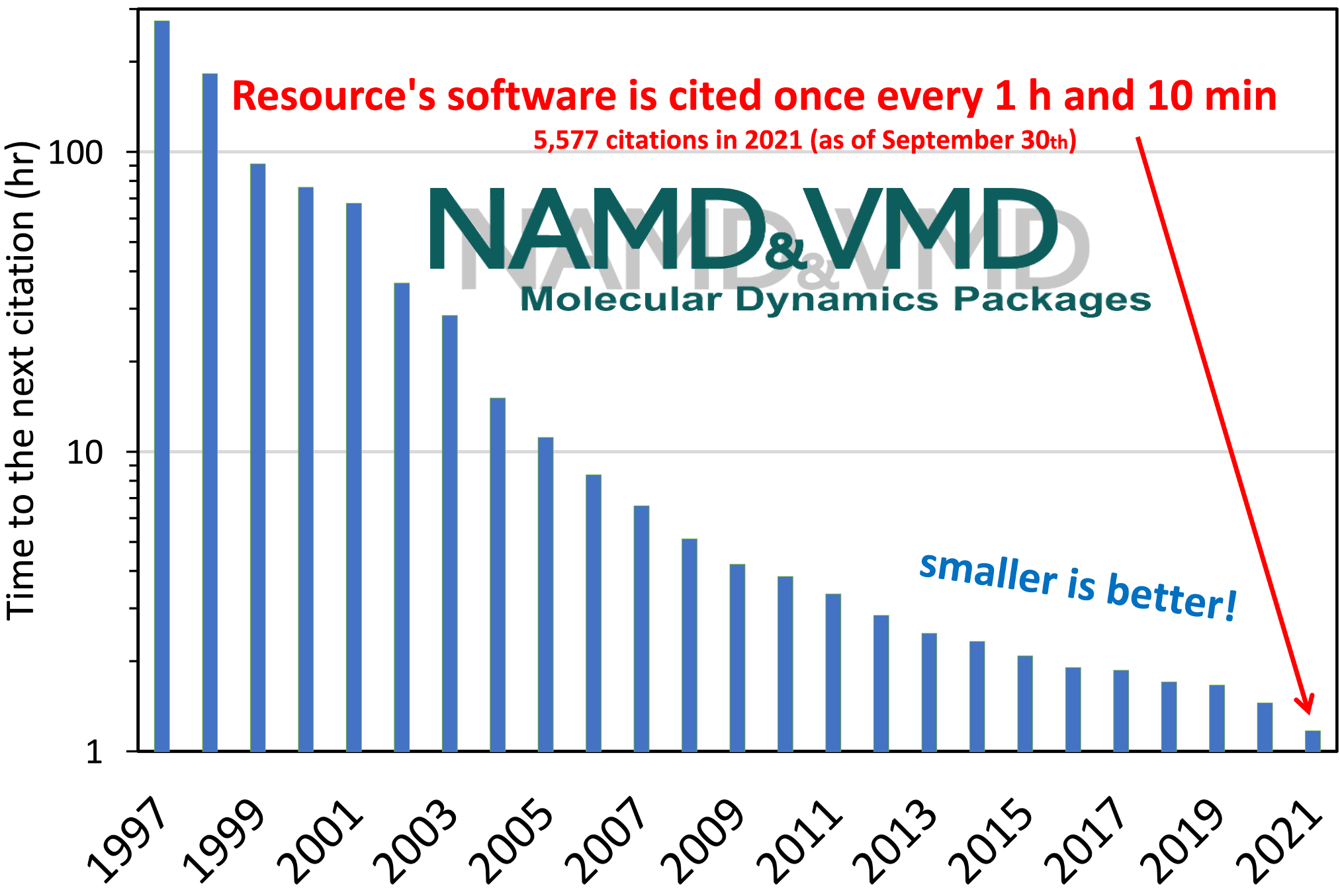
The computational microscope provided by the Resource's software programs,
NAMD and VMD,
has enabled hundreds of thousands of researchers worldwide
to make discoveries in molecular sciences.
While there is no simple metric to capture all the research
enabled by the Resource,
publications that have cited the Resource's software offer a solid measure
for the impact of the computational microscope.
Counting only the flagship programs VMD and NAMD,
the Resource continues to receive a staggering and increasing
number of citations every year,
arguing strongly for its high impact and substantial economies of scale.
Development and maintenance of these programs are supported by the NIGMS.
The human body produces its weight of ATP every day.
This universal currency of energy in biology is produced by the powerhouse of the cell, the mitochondrion.
It is then shipped into the cytoplasm of the cell to fuel a myriad of enzymes and biochemical processes
ranging from combustion of sugar molecules to neuronal excitation and muscle movement.
Interaction of the glycolytic enzyme hexokinase-II (HKII) with the mitochondrial ATP channel VDAC1
regulates the metabolism of the cell.
This interaction is amplified in many cancer cells, leading to faster glycolysis and cell growth and proliferation.
Simulations performed by the Center's software combining molecular dynamics NAMD and Brownian dynamics ARBD
show the central role of the membrane in binding of these proteins,
which in turn regulates mitochondrial permeability with direct implications in mitochondria-mediated cell death.
Read more in Communications Biology and Featured articles by TACC's Stampede2 HPC Supercomputers.
image size:
6.9MB
Made with VND
Modern neuroscience is undergoing a profound evolution due to systematic
efforts that provide large, comprehensive datasets on properties of
brain cells, their connectivity, and activity. Converting these widening
streams of complex data into new knowledge via analysis and modeling is
a major challenge for the field.
This rendering of a model of the mouse primary visual cortex (V1) with a
Neuropixels probe was awarded 2nd Prize in the 2021 NIH BRAIN Initiative
"Show
Us Your BRAINs!" Photo and Video Contest. The image was created with new
software created at the Center: Visual Neuronal Dynamics
— VND
— based on the Center's powerful VMD software, and
produced in collaboration with the Allen Institute.
The VND software performs visualization of bio-realisitc 3D
neuronal network models and aims to provide efficient
workflows for large and complex systems of thousands of neurons and
beyond. VND integrates the SONATA format, which
is used
by the Brain Modeling
Toolkit (BMTK), with the existing powerful visualization
capabilities of VMD to provide a platform for user-friendly, flexible,
and highly efficient visualization of bio-realistic neuronal network
models and simulations.
The initial version of VND reads SONATA file data, displays a new
neuroscience-specific interactive GUI, and creates neuronal network
visualizations for both exploratory visualization and for generating
publication-quality figures for research manuscripts.
(This image and VND were highighted in an August 2021 blog
post by NIH Director Francis Collins.)
Made with VMD
VMD supports the broadest possible range of hardware platforms of value to
molecular scientists. ARM processors have dominated mobile phone and
tablet platforms due to their energy efficiency and customizability. Our
investigation of GPU-accelerated ARM platforms for molecular modeling
demonstrated their potential, as
as we previously reported. Recently
ARM platforms have become significantly more performant, with today's
state-of-the-art 64-bit ARM CPUs beginning to turn up in top ranked
supercomputers, public clouds such as AWS, and both laptop and desktop
PCs. Apple has released new Mac laptop and desktop computers based on its
own "M1" 64-bit ARM processors. VMD 1.9.4 adds support for the new Apple
M1 platform and MacOS 11, a major new version of MacOS that supports
hardware based on both Intel x86 and Apple ARM M1 processors. The new VMD
builds for Apple M1 platforms uses 64-bit addressing to facilitate
efficiently working with very large macromolecular systems and
biomolecular complexes limited only by memory capacity. To provide the
best possible performance on the new ARM-based platforms by Apple and
other vendors, VMD incorporates hand-vectorized loops for NEON SIMD
instructions, enabling rapid alignment of atomic structures with cryo-EM
densities, and interactive animation of molecular surfaces with
QuickSurf, and molecular orbital visualizations from quantum chemistry
calculations and hybrid QM/MM simulations. The new VMD builds for Apple
M1 Mac hardware are available
from the VMD home page.