Highlights of our Work
2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
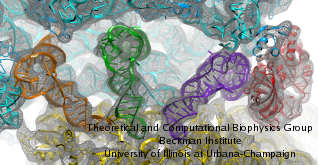
image size:
117.5KB
made with VMD
Living cells are brimming with activity, much of it due to their
manifold molecular machines pulling cargo, importing and exporting
molecules, digesting food molecules and transforming their energy,
reading and copying genetic messages, or synthesizing proteins from
these messages (the latter done by the ribosome). Static structures of
the molecular machines have been resolved through crystallography:
machines pressed into the confinement of crystals and frozen into
inactivity reveal their atomic level geometry through this methodology.
However, many machines, for example the ribosome, undergo large
conformational transitions during their cyclic action, but active
motions are hard to view in atomic detail. A way out is offered by
electron microscopy which freeze-shocks machines into states
characteristic for action cycle intermediates. Unfortunately, the method
does not yield atomic resolution images, leaving the chemical
detail needed for a comprehension of the mechanisms blurred.
Computational methods can be used to bridge the resolution gap: atomic level
structures of non-functional states of the machines captured in crystals
are deformed to match electron microscopy images. Until recently, the method
worked well only for very small machines. Now a team of electron
microscopists and computational biologists using NAMD extended the
method to common size machines and reported
its successful application to the ribosome, providing astonishing detail
about ribosome dynamics and function. For more details, see our MDFF website.



