Mechanosensing Spotlights
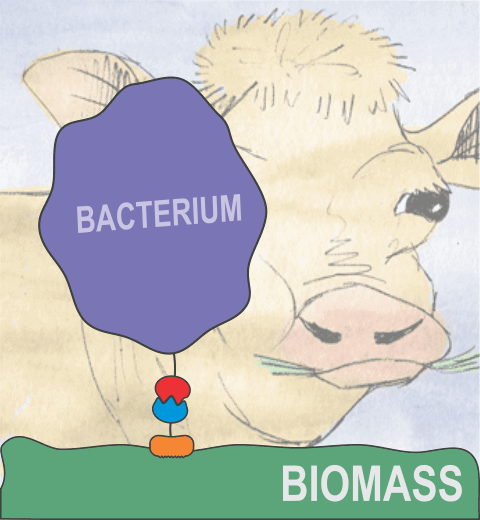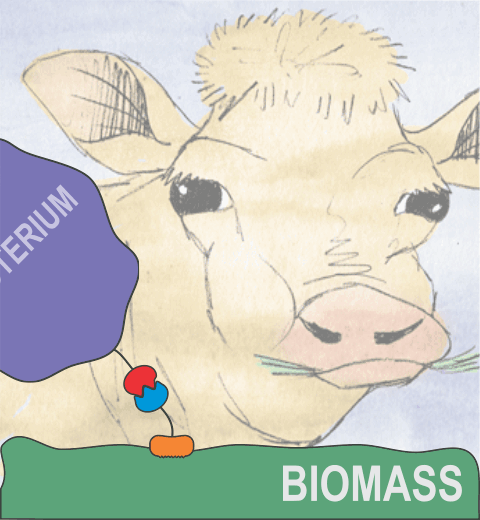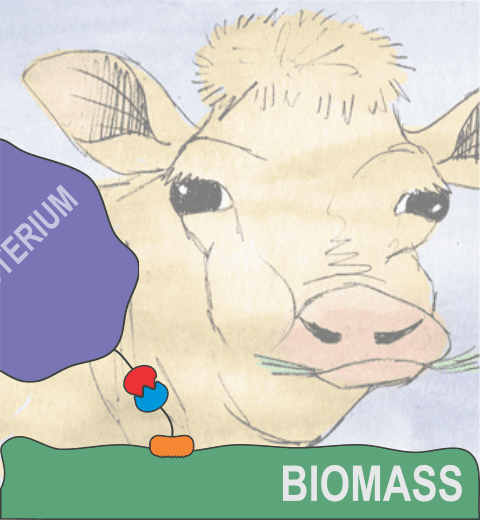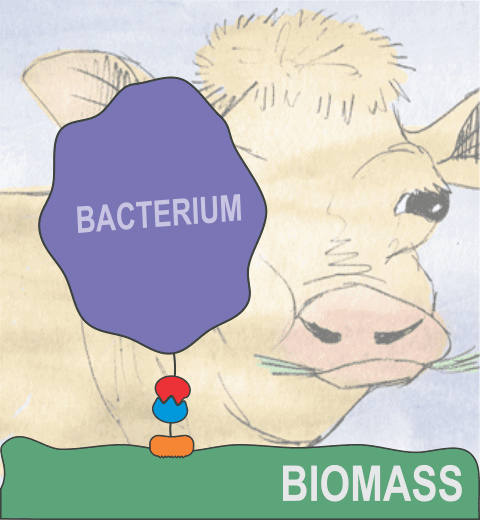
Bacteria can make a living from a very wide range of food sources. This ability makes them, for example, essential symbionts in animal digestive tracts where they assist their hosts in breaking cellulose fibers up into compounds degradable by the animal metabolism. Today, human gut bacteria, part of the human microbiome, are one of the hottest research topics in medicine. Gut bacteria face a particularly tough job in the rumen of the cow where they digest hardy cellulose fibers of grasses. Key to the job, taking place in a constantly moving fluid, are molecular tentacles, so-called cellulosomes, on the surface of the symbiotic bacteria. The cellulosomes develop a tight grasp on and then effective cleavage of cellulose. In a joint experimental-computational study researchers have investigated how in case of the bacterium Ruminococcus flavefaciens cellulosomes are built in a modular way, with molecular modules easily binding and unbinding during cellulosome construction, but sticking extremely strongly together during cellulosome digestive activity. As reported recently, single molecule force microscopy and molecular dynamics simulations using NAMD could show that under strain the adhesive bonds between cellulosome modules become stronger than seen in any other biomolecular system, in fact, become nearly as tight as strong chemical bonds. While the experimental data revealed bond strength and other characteristics, simulations reproducing the observed data provided a detailed view of the adhesive bond at atomic resolution, thereby revealing the physical mechanism underlying the uniquely adhesive property of cellulosomes. Gut bacteria and cellulosomes can be employed in 2nd generation biofuel generation (see highlight Waste into Fuel). More on gut bacteria and cellulosomes on our biofuels website.

image size:
110.0KB
made with VMD
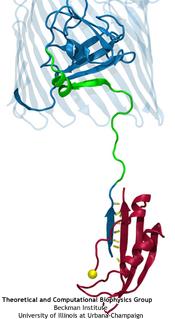
The human body is protected by self-healing mechanisms, one of them being instant blood clotting at a bleeding site after blood vessel injury. What triggers the formation of a blood clot? Researchers found that a protein on blood platelets, called GPIbα, functions as a sensor of so-called high shear flow caused by bleeding. A loop-shaped, 17-amino-acid-long, segment of GPIb&alpha, the β-switch, acts as the flow sensor. Once a blood vessel is injured, bleeding increases shear stress due to blood flow at the wound, which in turn induces the β-switch to change from a loose, loop-shape to an elongated, hairpin-shape, the latter referred to by researchers as a β-hairpin. This conformational change makes GPIbα stick better to the damaged vessel and eventually leads to blood clotting, which heals the vessel. In a prior study (see the Jul 2008 highlight, Molecular Flow Sensor Triggers Wound Healing), Molecular dynamics simulations using NAMD and VMD provided already a microscopic view of the flow-induced loop to β-hairpin transition. A recent study extended the investigation of the remarkable biological flow sensor, detailing the flow rate needed to trigger it and identifying the detailed sensor mechanism. A combination of simulation and mathematical analysis revealed the β-switch as a system of two stable states, one disordered, with loop geometry and one ordered, with β-hairpin geometry. Normal flow prefers the disordered state; high shear flow prefers the ordered state, inducing thereby the life saving transition. More on our flow sensor website.
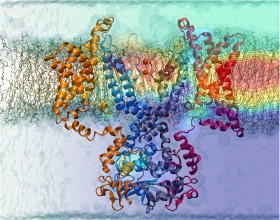
Nerve cells, through their electrical signals, control actions and intelligence of higher organisms. The signals result mainly from potassium and sodium ion channels in the cells: when the cells are stimulated electrically, they send an all (in case of sufficient stimulation) or nothing signal to other nerve cells or organs like muscle. As shown in ground breaking work by 1963 Nobelists Hodgkin and Huxley, cast into mathematical equations, nerve cells establish these signals through voltage gating of channels. The nature of the gating, monitored through the so-called gating current, has been elusive for decades, despite a detailed characterization of the ion conductivity itself rewarded through a 2003 Nobel Prize to MacKinnon. The riddle is that the channel involves a protein with few charged amino acids that seem to be only weakly coupled energetically to an electrical potential gradient across the cell membrane. Now a sweeping modeling study using NAMD employing the most powerful computers available to researchers today has led to an explanation of voltage-gating. Simulations revealed that the potential gradient is focused by the channel protein to a very narrow region such that its value is much larger than anticipated. The protein was also seen to arrange its charged amino acids sensing the gradient in an unusual helix, a so-called 310 helix, that aligns charges perfectly while at the same time inducing a motion that opens and closes the channel. Proof of the veracity of the computational model is that the calculated gating current fits perfectly the observation. More on our potassium channel website.
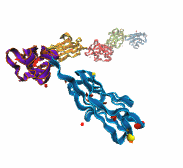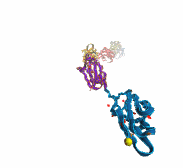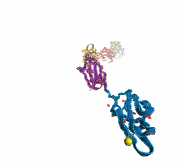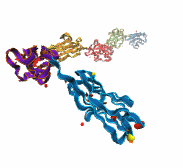
image size:
212.3KB
made with VMD
A smart strategy usually involves a plan B. As it turns out, the muscle proteins in our bodies responsible for the physical motions like running or the beating of our hearts, also rely in their function on having a plan B strategy. When contracting and extending, muscle fibers generate tremendous forces that need to be buffered to protect muscle from damage. This role falls to the muscle protein titin, which is composed of a chain of linked domains, making it a molecular rubber band. When a small force is applied, titin employs its plan A and stretches apart without unraveling its individual domains (like what the movie on the side shows). When a stronger force is applied, plan B kicks in and further elasticity is generated by the unwinding of the protein domains one at a time. By practicing two modes of response to different levels of forces, titin provides the elasticity that muscle needs at a minimal structural cost. A recent computational-theoretical investigation has provided a molecular view on how titin's two plans work, the study featured in a journal cover. The needed simulations were performed using NAMD. Principles described in this study can also be found in other mechanical proteins, recently reviewed here. More on our titin IG6 website.
Viruses are the simplest life forms known. In fact, one can question if they are life forms at all, as they cannot exist without infecting a host cell and using its machinery for replication. The virus is indeed just a package material surrounding a genetic message that instructs the host cell to replicate the virus. It looks like a soccer ball, but is a million times smaller (see also the March 2006 and January 2007 highlights). The infection, a well known example being infection of human cells by a flu virus, involves the virus to approach a human cell and dock onto it, become internalized by the cell, bursting then its package, called the capsid, and release the genetic message. The virus capsid needs to be sturdy and impermeable up to the approach to the cell, but then become brittle and porous to release the genetic material. Obviously, the virus capsid must have very distinct mechanical properties to function. To investigate these properties experimental and computational biophysicists teamed up. The experimentalists placed empty capsids of the hepatitis B virus onto a small chip and mechanically squeezed the capsid then with an extremely small tip, measuring how much force is needed to squeeze the spherical capsid down repeatedly. Computational researchers using NAMD repeated the experiment in simulation. As they reported recently, simulation gave the same forces as the experiment, but yielded also a detailed picture of the capsid mechanics. More on our "Molecular dynamics of viruses" web site.

image size:
661.9KB
made with VMD
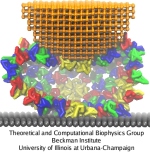
Biological cells are surrounded by a highly versatile, yet very feeble cellular membrane and need to balance differences between the cell's interior and exterior that otherwise would burst the membrane. For example, the osmotic pressures inside and outside the cell need to be closely balanced. Thousands of proteins in the membrane act as gatekeepers, opening pores that can also act as safety valves, helping to reduce the interior-exterior difference in pressure rapidly. One such protein, the mechanosensitive channel of small conductance MscS (see the Mar 2008 highlight, "Observation and Simulation Depict Cell's Safety Valve", the Feb 2007 highlight, "Observing and Modeling a Crucial Membrane Channel", the May 2006 highlight, "Electrical Safety Valve", and the Nov 2004 highlight, "Japanese Lantern Protein") opens in response to cellular membrane tension generated due to a drastic imbalance in osmotic pressure as it arises when a bacterial cell suddenly finds itself in fresh water, rather than a highly saline physiological medium. The MscS channel widens then to jettison molecules out of the cell and reduce tension on the cellular membrane quickly. In a recent report, researchers have combined experimental data from electron paramagnetic measurements and computer modeling to reveal in atomic detail how MscS opens and closes its channel. Combining measurement and modeling, the researchers established a highly resolving computational microscope, unmatched by existing microscopes (more on our MscS website).
image size:
97.1KB
made with VMD
Adhesion between human cells organizes our body into its organs and parts. The adhesion comes about through an intricate system of molecules that perform their task in a highly selective manner such that the body's different types of cells will find the right cells and stick to them. This selectivity leads to tissue differentiation and the organization of organs as complicated as the brain. Cadherin proteins form a particular family of such adhesion molecules. Interestingly, they glue cells together only in the presence of calcium. Some members of the cadherin family of proteins are also involved in the transduction of sound and cadherin mutants are known to cause hereditary deafness (see the April 2005 highlight, "Hearing: Turning Sound into Voltage"). How cadherins selectively bind to each other and the role of calcium was not well understood, but now molecular dynamics simulations have offered magnificent insight into calcium's role as recently reported. The simulations took advantage of parallel supercomputers and NAMD's ability to harness their power. The simulations revealed that in the absence of calcium cadherins stick out of cell surfaces like ends of loose rope; in the presence of calcium cadherin molecules turn into stiff hooks that link cells together. The calcium-induced links can withstand the strong mechanical forces that arise between cells much larger than cadherin (more on our cadherin website).
Like all organisms, bacteria have to eat. However, bringing nutrients in from the outside world is not an easy task for many bacteria that are surrounded by an extra membrane. The second membrane, called the outer membrane, offers additional protection but at a cost: no energy can be generated or stored at the outer fringes of the cell. So, to import large, rare nutrients that cannot cross by diffusion alone, bacteria have evolved a unique transport system which couples the inner, energy-generating membrane to the passive outer membrane, known as the TonB-dependent transport system. TonB, an inner membrane-associated protein, transfers energy across the periplasm to a variety of outer-membrane transporters. These transporters have a large, beta-barrel structure which is blocked in the middle by a plug called the 'luminal domain'. How TonB transfers energy to the transporter and causes the luminal domain to come out is still a mystery though. Now with the help of computer simulations using NAMD and a recent crystal structure of TonB coupled to BtuB, the transporter responsible for vitamin B12 transport, researchers have shown that TonB can mechanically activate the transporter by pulling on the luminal domain, causing it to leave the barrel. Using steered molecular dynamics, it was found that TonB stayed firmly attached to the luminal domain of BtuB, even though the contact between the two is limited to just a handful of residues. Furthermore, this pulling initiated unfolding of the luminal domain, opening a transport pathway for the substrate. These results, the subject of a recent publication and also highlighted in Science, demonstrate how a mechanical coupling can bridge the gap between the two membranes, thus enabling outer membrane transport.

image size:
91.7KB
movie (
789.9KB
)
Mechanical forces are everywhere in human life. Strong forces power machines and cars, our body's forces let us labor and move, soft forces are sensed through touch, even softer ones through hearing. Forces are also ubiquitous in the living cell, driving its molecular machines and motors as well as signaling ongoing action in its surroundings. Man made, force bearing machines rely on extremely strong materials not found in the cell. How can the cell bear substantial forces? Also, how do cells sense extremely weak forces as in hearing, surpassing most microphones? Single molecule measurements, reviewed in a recent issue of Science, begin to answer these questions offering information on biomolecules' mechanical responses and action. However, the information offered by these measurements is not enough to relate the biomolecular function to the biomolecular architecture. Biomolecules in cells can move in amazing ways, but we did not know why. As one contribution in the Science issue demonstrates, computational modeling comes to the rescue. It can simulate the measurements and, in doing so, can reveal the physical mechanisms underlying cellular mechanics at the atomic level. In as far as observed data are available, the simulations show impressive agreement with actual measurements. While initially only following experiments or, at best, guiding experiments, modeling has advanced now further and through simulated measurements discovered on its own entirely novel mechanical properties that were later verified by experimental measurements. Experimentalists reacted to the new competition and began to do simulations themselves. More here.
Mammalian cells adhere to each other forming tissues. The adhesion is due to a network of proteins, so-called extracellular matrix proteins, "gluing" the cells together. The cell membranes are too soft to provide anchoring points for the extracellular matrix proteins; rather, the cells furnish on their outer surface specialized hooks for anchoring the extracellular matrix proteins. The hooks, in the form of surface proteins, are linked directly through the membranes to the intracellular cytoskeleton that stabilizes and shapes cells. Integrins are an important family of such surface proteins that form hooks specific for certain types of extracellular matrix proteins. The hooks are flexible, they can be open for contacts or closed, the switch being induced by signals from inside or outside the cell through interactions with other proteins. The interactions between integrins and extracellular matrix proteins are rather complex, as the proteins are composed of many subunits; fortunately, their overall structures are presently being solved through crystallography. In a recent report a major component of an integrin and an extracellular matrix protein have been investigated through molecular modeling using NAMD, including steered molecular dynamics. The study described in detail how the extracellular matrix protein induces a transition in integrin, potentially strengthening its adhesion property. See also previous highlights: the May 2006 "Killer's Entry Route", Dec 2004 "Snap Fastener on Biological Cells", Dec 2003 "Body's Glue", and Mar 2002 "Cells Sense Push and Pull". More on modeling of extracellular matrix proteins and integrins can be found here.
image size:
300.4KB
made with VMD
The living state of biological cells manifests itself through mechanical motion on many length scales. Behind this motion are processes that generate and transform mechanical forces of various types. As with other cell functions, the machinery for cellular mechanics involves proteins. Their flexible structures can be deformed and restored, and are often essential for handling, transforming, and using mechanical force. For instance, proteins of muscle and the extracellular matrix exhibit salient elasticity upon stretching, mechanosensory proteins transduce weak mechanical stimuli into electrical signals, and so-called regulatory proteins force DNA into loops controlling, thereby, gene expression. In a recent review, the structure-function relationship of four protein complexes with well defined and representative mechanical functions has been described. The first protein system reviewed is titin, a protein that confers passive elasticity on muscle. The second system reviewed is the elastic extracellular matrix protein fibronectin and its cellular receptor integrin. The third protein system covered are the proteins cadherin and ankyrin involved in the transduction apparatus of mechanical senses and hearing. The last system surveyed is the lac repressor, a protein which regulates gene expression by looping DNA. In each case, molecular dynamics simulations using NAMD provided insights into the physical mechanisms underlying the associated mechanical functions of living cells. (more on our mechanobiology web site).
image size:
301.5KB
made with VMD
Muscle fibers, through their so-called thick and thin filaments, contract and extend in doing their work. To render the fibers elastic and protect them from overstretching, the thick filaments are connected through a long and thin elastic protein, titin, to the base of the fibers. Titin, by far the longest protein in human cells, is a molecular bungee cord and, like such cord, must be affixed firmly to the base. How this is done was a mystery until crystallographers took the first atomic resolution image of the system: it turns out that two titins are spliced together at their ends like ropes. The splicing involves a third small protein, the titin-telethonin-titin system forming a U. The U apparently is thrown over a bollard-like cellular structure to hold the thick filaments much like boats are held by bollards and ropes at their mooring place. The crystallographers teamed up with computational biologists to investigate the mechanical strength of the titin - telethonin - titin cord by means of molecular dynamics simulations using NAMD. As reported recently, the cord has great mechanical strength due to an extended network of hydrogen bonds between beta-strands, common structural features in proteins, that in the present case form a sheet extending through all three proteins. This discovery explains how living cells can splice cellular proteins together through a system of hydrogen-bonded beta-strands that extend through several proteins. Interestingly, such beta-strands were seen previously in cases of diseases like Alzheimers where the feature leads, however, to pathological assembly of proteins. What needs to be understood now is how the telethonin glue is applied only to the right spots in the cell and how the cells prevent telethonin from splicing together the wrong proteins. For more information visit our titin-telethonin web page.
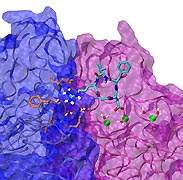
image size:
198.4KB
made with VMD
Biological cells must be capable of attaching themselves to their surroundings. For this purpose they utilize fibrillar proteins, such as fibronectins, that grasp cells through cell surface receptors integrins. The latter act as snap fasteners to the extra-cellular fibrils. The growth, movement, and survival of cells are all dependent on the ability of integrins to fasten cells upon intra-cellular signals or to signal inwards that something has become fastened on the cell surface. The major fastener on integrins are simple divalent ions like Mg++ or Ca++ that can adhere to specific molecules with amazing strength, even though the interaction at the cell surface is exposed to water. Computer simulations using NAMD, reported recently, revealed a dynamic picture of the interactions used by cells to link themselves to the extra-cellular matrix. They showed that it is actually a brave water molecule that is recruited by integrins as a protective shield for the interaction. The simulations provide for the first time a detailed view of how cell tissues are stabilized through surface ions against mechanical stress.
image size:
170.9KB
made with VMD
One of life's great achievements is the development and maintenance of multi-cellular organisms, from an embryo to adulthood. Multitudes of cells need to be sorted and resorted into tissues, organs, and body of living beings. One strategy towards this end is to endow cells with so-called adhesion proteins that connect a mechanical framework inside cells through the cell membrane with other cells. A key type of adhesion protein is cadherin (calcium-dependent adherent protein) that stretches through the cell surface five-tandem domains. The outermost domain can stick to a cadherin molecule from an adjacent cell. Crystallography provided the molecular structures of cadherin pairs and resolved in atomic detail the cadherin-cadherin contact between cells. This prompted a collaboration that aimed at probing the adhesion strength of cadherin pairs through steered molecular dynamics simulations stretching the pairs apart. Results of the simulations were reported in a recent publication that employed NAMD as well as VMD. As shown by crystallography, the cadherins each insert a tryptophan residue into the other protein. The link thereby established can be broken only through strong forces that induce a step-wise slippage of the residues first out of their binding pockets and then along the protein surface. This scenario suggests a mechanism for selectivity among cadherins, i.e., why among the various cadherins found on the surfaces of cells some adhere much better to each other than others.
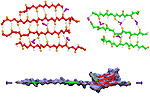
image size:
246.4KB
Movies of stretching FN-III-1:
real media
(
119.5KB
)
windows media
(
125.9KB
)
mpeg (high quality)
(
2.2MB
)
Image and movie made with VMD
Tissues of the human body are composed of specialized cells held together by a connective fabric of proteins, that form the knots of a net glueing cells together. Upon stretching tissues, the knots unravel, rendering the net larger, but mysteriously also firmer. A protein called fibronectin-III-1 plays a particularly important role in the latter respect. Atomic force microscopy revealed that under mechanical tension fibronectin-III-1 stretches to ten times its initial length; but is does so in two steps, the first stretching step leading to net strengthening. It had been discovered earlier that other fibronectins found between cells are made of two sheets packed like a sandwich, but the structure of fibronectin-III-1 remained elusive. In an experimental-computational collaboration reported recently, the structure has now been resolved that at first sight looked similar to the sandwich structure of the other fibronectins, but on closer inspection showed a weak and a strong sheet. Simulations using NAMD revealed that stretching of the protein unravels readily the weak sheet, and only therafter the strong sheet. It turns out that the strong sheet of fibronectin-III-1 by itself, known as anastellin, inhibits tumor growth. Stretching of fibronectin-III-1, as it occurs naturally in tissue, unravels apparently half of the protein to render it extremely adhesive, strengthening a protein net that prevents metastasis of cancer cells and also assists wound healing (press release, more).
Living cells sense their environment and respond to its changes through proteins integrated into their outer membrane. These proteins mediate a broad range of cellular activities, including active and passive transport of materials across the membrane as well as response to osmotic shock, which can strain the cell membrane to the point of catastrophic bursting. Cells protect themselves through so-called mechanosensitive channels that open before the membrane tension grows too large. Molecular dynamics simulations and advanced analysis using NAMD and VMD have revealed in a recent report how the joint mechanics of membrane and protein opens a mechanosensitive channel called MscL. The finding promises to revolutionize the modern view of membrane - protein interaction: the membrane, far from being a homogeneous elastic sheet, exhibits a dramatic variation of tension across its thickness that proves to be decisive for the opening of MscL. More here.
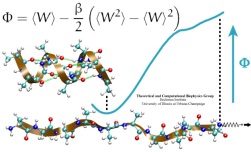
image size:
76.6KB
made with VMD
Many proteins in living cells are nanomachines that undergo mechanical
transformations. Modern modeling methods permit the manipulation of
such proteins to discover the physical mechanism behind their function.
Applying forces, one can induce geometrical changes characteristic of the
proteins' role in the cell and, beyond obtaining qualitative insight,
calculate the work
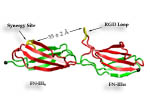
image size:
85.1KB
made with VMD
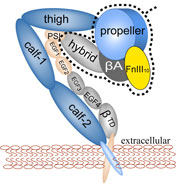
Cells in animals adhere to dynamic, seemingly random assemblies with other cells that make up tissues like skin, organs, and brain. The cell's adhesion and motion is controlled by the extracellular matrix, with the protein fibronectin being a key component. The proteins have optimal mechanical elasticity and also signal to cell surface receptors, integrins, the tension exerted on them. How this is achieved is the subject of an ongoing collaboration with the research group of Viola Vogel of the Department of Bioengineering at the U. of Washington in Seattle (see also Oct 2001 highlight). The most recent publication from this effort reports a 97,884 atom steered molecular dynamics simulation using NAMD. It is revealed now that stretching two consecutive domains of fibronectin deforms two sites, the so-called RGD and synergy sites as well as their distance. This weakens binding to cell receptors and, as a result, integrins can function as gauges that signal the magnitude of exterior forces to a cell.
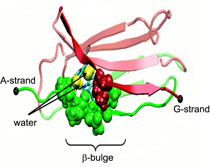
image size:
78.8KB
made with VMD
Mechanical force is seen today as a key component of molecular processes in cells: forces can be signals as in touch receptors, products as in muscle action, and substrates as in the matrix surrounding moving cells . This view of molecular processes is the result of a series of ground-breaking investigations that have become possible only recently and is rapidly turning into a new field, mechanobiology (see recent review). A collaboration with V. Vogel and coworker (U. Washington, Seattle) has investigated structural changes accompanying stretch-induced unfolding events in type III fibronectin, a protein of the extracellular matrix, explaining the design of these proteins.
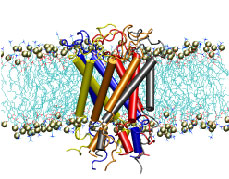
image size:
247.9KB
made with VMD
"How do you feel?" Biologists now have an answer that may surprise you. Our sense of touch relies upon the fact that cells in our fingertips can sense the pressure from a tabletop and transmit a signal to the brain. But until recently, the molecular mechanism for turning the stretching of a cell membrane into a cellular signal was unknown. An important step in understanding this process was the discovery of a protein known as a the mechanosensitive channel of large conductance, or MscL. Though this protein has been studied primarily in bacteria, homologues exist in all major kingdoms of life. Researchers in the Theoretical Biophysics Group have used molecular dynamics simulations to study, at the atomic level, how MscL opens in response to pressure changes. Models of MscL will give us new insight, not only into how we feel, but also how our hearts beat and how we keep our balance. Feel better now? ( more, publication )

image size:
106.6KB
made with VMD
Water can act as a conformational lubricant for protein folding. The giant muscle protein titin is a roughly 30,000 amino acid long filament which plays a number of important roles in contraction and elasticity. For example, upon stretching in muscle some of titin's protein domains can unfold one-by-one permitting titin to retain elastic properties in muscle over a very wide range of length. To examine in atomic detail the dynamics and structure-function relationships of this behavior, SMD simulations of force-induced titin domain unfolding were performed in close collaboration with atomic force microscopy observations. The simulations led to the discovery that water molecules play an essential role in breaking sets of hydrogen bonds that control the unfolding of titin's domains (see resulting publication).