Computational Force Microscopy
Computational Force Microscope
 Atomic vs Computational force microscopy. The protein being stretched is ankyrin, likely involved in many mechanical processes of living cells;
ankyrin repeats are thought to form part of the transduction apparatus in hearing.
Atomic vs Computational force microscopy. The protein being stretched is ankyrin, likely involved in many mechanical processes of living cells;
ankyrin repeats are thought to form part of the transduction apparatus in hearing.
Mechanical forces are everywhere in human life. Strong forces power machines and cars, our body's forces let us labor and move, soft forces are sensed through touch, even softer ones through hearing. Forces are also ubiquitous in the living cell, driving its molecular machines and motors as well as signaling ongoing action in its surroundings. Man made, force bearing machines rely on extremely strong materials not found in the cell. How can the cell bear substantial forces? Also, how do cells sense extremely weak forces as in hearing, surpassing most microphones? Single molecule measurements begin to answer these questions offering information on biomolecules' mechanical responses and action. However, the information offered by these measurements is not enough to relate the biomolecular function to the biomolecular architecture. Biomolecules in cells can move in amazing ways, but we did not know why. As a review in Science demonstrates, computational modeling comes to the rescue. It can simulate the measurements and, in doing so, can reveal the physical mechanisms underlying cellular mechanics at the atomic level. In as far as observed data are available, the simulations show impressive agreement with actual measurements. While initially only following experiments or, at best, guiding experiments, modeling has advanced now further and through simulated measurements discovered on its own entirely novel mechanical properties that were later verified by experimental measurements. Experimentalists reacted to the new competition and began to do simulations themselves.
Steered Molecular Dynamics
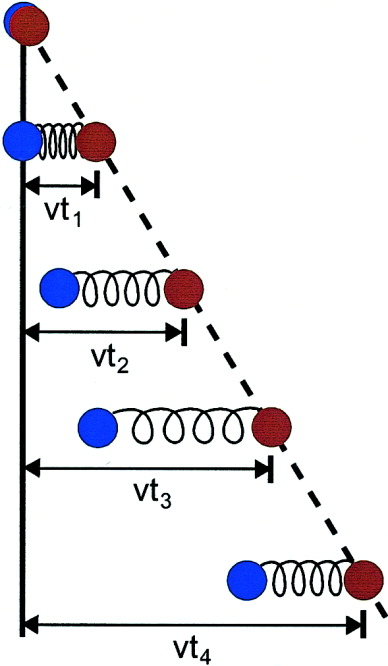 Constant velocity SMD simulation.
Constant velocity SMD simulation. The equilibrium dynamics of a biomolecule at the atomic level can be readily observed using molecular dynamics simulations. Even more can be learned about the architecture and function of a biomolecule by using simulations in which external forces are applied to it. These kind of simulations, called steered molecular dynamics (SMD) simulations, were first performed a decade ago following experiments in which atomic force microscopy experiments recorded the unbinding forces of gluing biomaterials at the single molecule level. Steered molecular dynamics has evolved and is now a standard capability of NAMD, featuring two typical protocols. In the first protocol, a constant force is applied to one or more atoms; extension or displacement is then monitored throughout dynamics. Customized time-dependent forces may be applied as well. In the second protocol, called constant velocity SMD, a harmonic potential (spring) is used to induce motion along a reaction coordinate (see Figure). The free end of the spring is moved at constant velocity, while the protein atoms attached to the other end of the spring are subject to the steering force. The force applied is determined by the extension of the spring and can be monitored throughout the entire simulation. SMD has become now a standard tool complementing single molecule experiments in vitro.
- Click here for a movie (mpeg, 4.1M) showing an SMD simulation in which ankyrin repeats are first stretched (tertiary structure elasticity) and then unfolded (secondary structure elasticity). Tertiary and secondary structure elasticity of ankyrin repeats have been observed through AFM experiments as well.
- Click here for a movie (mpeg, 2.8M) showing an SMD simulation in which ankyrin repeats are unfolded (secondary structure elasticity). The stepwise unfolding of ankyrin repeats observed in this simulation was later on confirmed through AFM experiments.
Reviews on Steered Molecular Dynamics
Recent Applications of Steered Molecular Dynamics (2006-2008)
Prior Applications of Steered Molecular Dynamics
Related TCB Group Projects
- Mechanical Proteins
- Cells Get Sticky with Calcium
- Molecular Basis of Fibrin Clot Elasticity
- Molecular Basis of Hearing
- Multiscale Modeling of Gene Repression
- Mechanical Strength of the Titin/Telethonin Complex
- Fibronectin and Integrin
- Unfolding Titin Immunoglobulin Domains
- Molecular Motor Scooting along DNA
- Bacteria Swim and Tumble
- IMD and the Glycerol Channel
- Unbinding of Retinoic Acid from its Receptor
- Molecular Basis for Anthrax Intoxication
- Mechanosensitive Channel MscS
- Mechanosensitive Channel MscL
Page created and maintained by Marcos Sotomayor.



