Second-Generation Biofuels
|
|
|
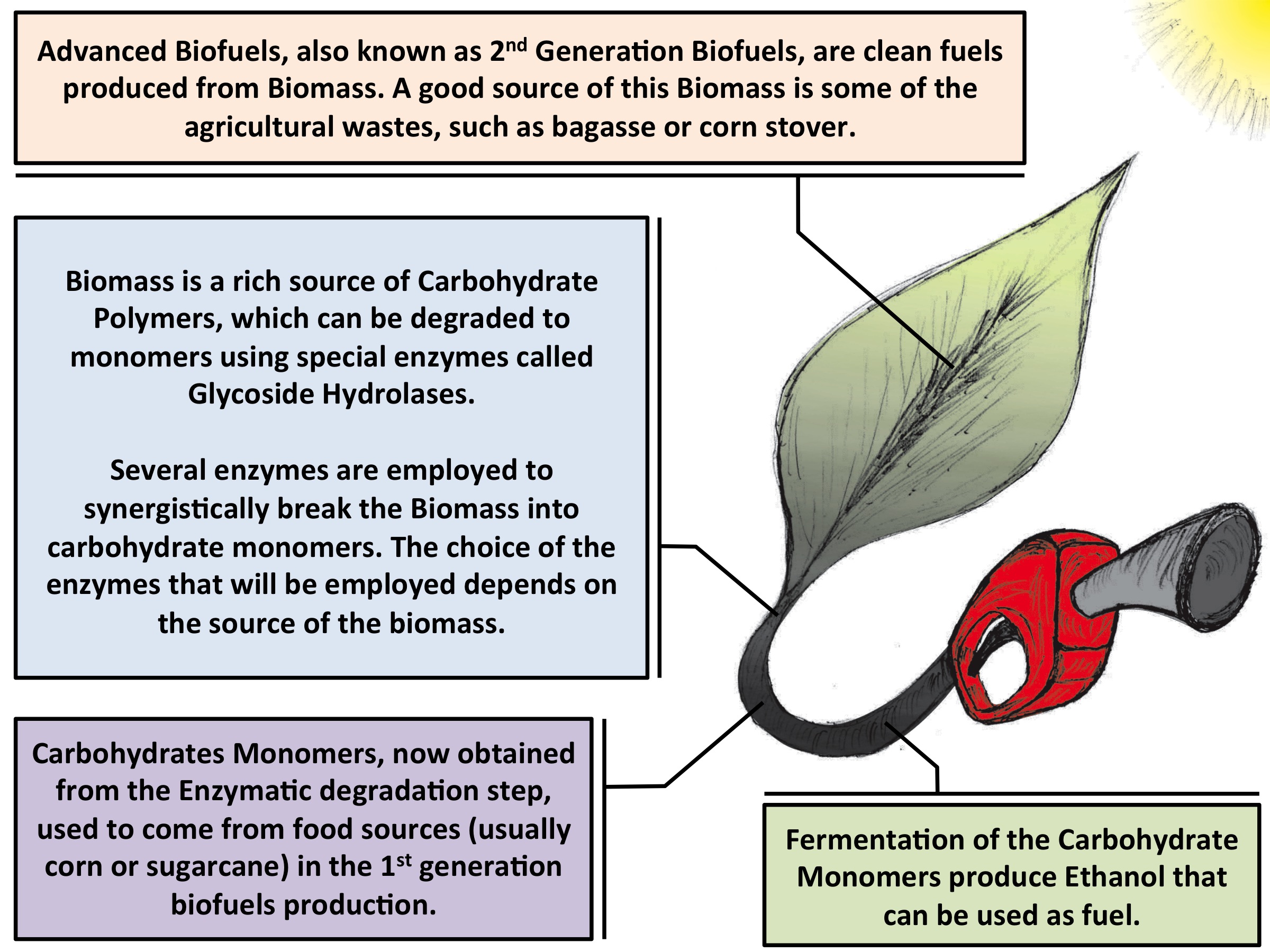
Fig. 1 - Illustration of the second-generation biofuel production process. The development of more efficient enzymes is a key point of where molecular dynamics can contribute. |
Overview
Greenhouse gas accumulation in the atmosphere, caused by the large consumption of fossil-derived fuels, coupled with the reduction of the world's supply of these fuels, has lead to increased interest in identifying novel sources of energy. Biofuels, produced from crops such as corn, represent an alternative energy source, however, their competition with food production results in an ethical dilemma. Fortunately, a solution is offered by second-generation biofuels, which can be produced from plant cell wall polysaccharides derived from agricultural waste, or biomass. Over the years, several technologies have been developed for the conversion of biomass to fermentable sugars, and future efforts aim to make this process cost-competitive in the present-day market.
Biofuels Production Problems
|
|
|
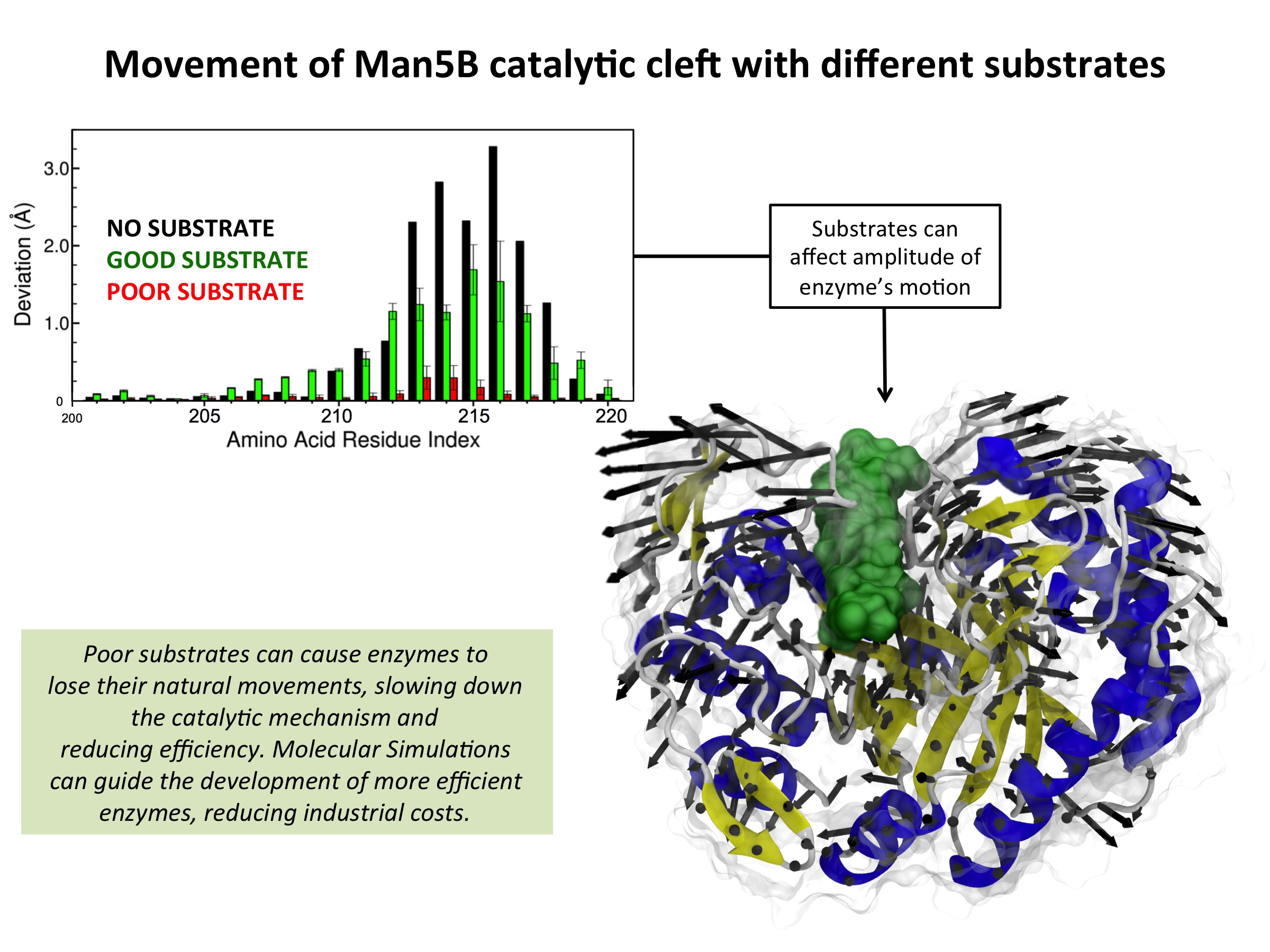
Fig. 2 - Illustration of the Man5B inhibition mechanism. This mechanism was suggested based on Molecular Dynamics Simulations with NAMD together with experimental data. Read more |
Through molecular dynamics study of Caldanaerobius polysaccharolyticus Man5B, an industrially important enzyme that cleaves both β-1,4 glucosidic and β-1,4 mannosidic linkages in biomass, we identified the mechanism of unsuitable substrate inhibition. Representative cleavage products cellohexose and mannohexose were docked into the Man5B catalytic site, and simulations of these model complexes indicated a notable loss of enzyme flexibility for the gluco-oligosaccharide cellohexose, as compared to the manno-oligosaccharide mannohexose. These results, combined with evidence from previous experimental studies, suggested that reduction of catalytic efficiency is related to the loss of enzyme flexibility in the presence of gluco-oligosaccharides, which serves to inhibit binding of new suitable substrates. Manno-oligosacchardies do not constrain enzyme flexibility, and thus do not inhibit substrate binding. Our conclusions are supported by kinetic experiments that demonstrate gluco-oligosacharides take much longer to exit the catalytic pocket, thus preventing new substrates from reaching the active site.
From Single Enzymes to Cellulosomes
|
|
|
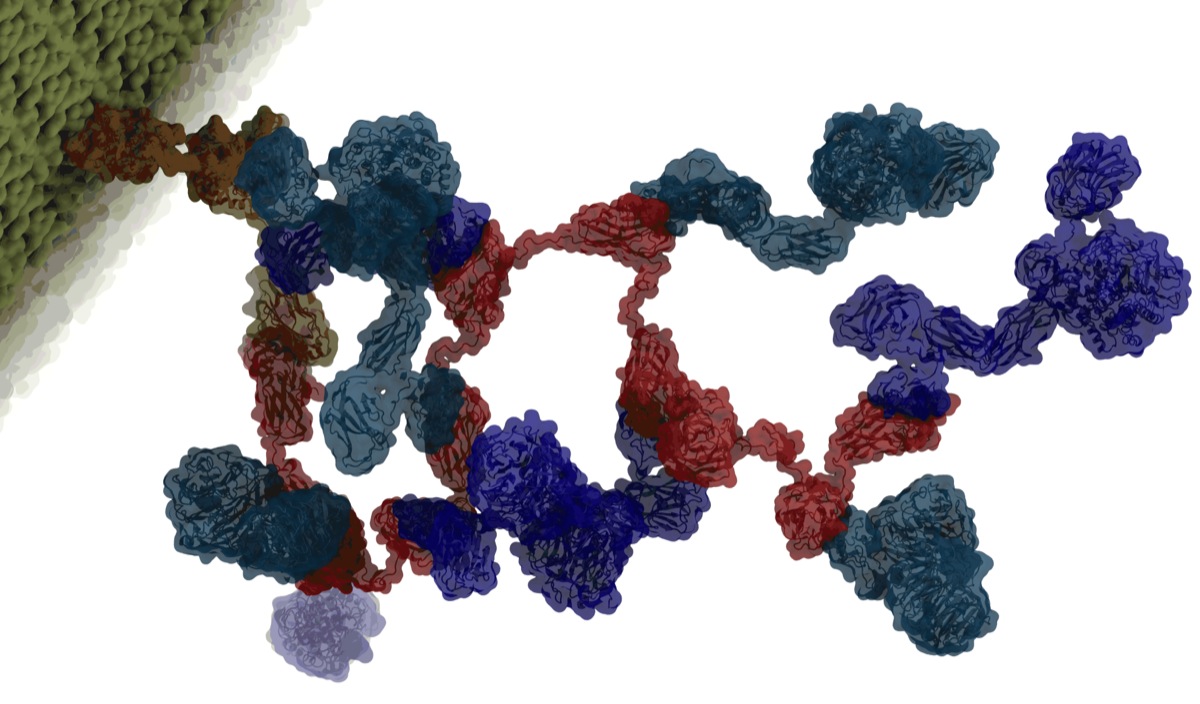
Fig. 3 - Illustration of the Cellulosome structure. |
Some bacteria employ a biomass degradation strategy based on large enzymatic complexes, which incorporate multiple enzymes working synergistically toward catalytic processing. These complexes, called cellulosomes, offer a promising approach to reduce biofuel production costs. Further, because cellulosomes are typically found in harsh natural environments, they have evolved ultra-stable structural characteristics that suggest novel biotechnological applications.
Cellulosomes represent extracellular proteinaceous assemblages comprised of a scaffolding backbone, onto which cohesion-containing modules for carbohydrate binding and catalytic activity are appended. Because the various modules exhibit a plethora of binding and enzymatic activities that facilitate cleavage of plant cell wall polysaccharides, the cellulosome serves as a virtual "Swiss army knife" for biomass degradation.
It is reported that Clostridium thermocellum, the most studied cellulosomal organism, has one of the highest rates of cellulose utilization known in nature. It's cellulosomal system displays a specific activity against crystalline cellulose that is 50-fold higher than the analogous non-cellulosomal system in the fungus Trichoderma reesei.
In order to better understand the efficient biomass degradation mechanisms of cellulosomal machineries, as well as their unique structural integrity, it is first necessary to study the interactions and attachment modes of the various subunits, and their process of synergistic action. We are currently collaborating with experimentalists in USA, Germany, and Israel to unravel the structure and catalytic functions of cellulosomes.
Sampling Conformations of Cellulsome Domains
Cellulosomes are formed by interactions of dockerins (Doc) and cohesins (Coh) that are connected by flexible linkers made of proteins with just a few to more than a thousand amino acids. The many flexible linker regions allow for very complex structural dynamics.
Some insight into cellulosome dynamics can be gained by determining possible conformations through the use of enhanced sampling methods. GSAFold, the implementation of Generalized Simulated Annealing (GSA) in NAMD, is employed here to explore conformations of the linker region. The structure used in the present simulation is from the Protein Data Bank (ID code 3KCP). The stochastic calculation with GSAFold, where only the linker region was allowed to rotate show that the system is most likely to be found in two conformations, see Figure 4.
|
|
|
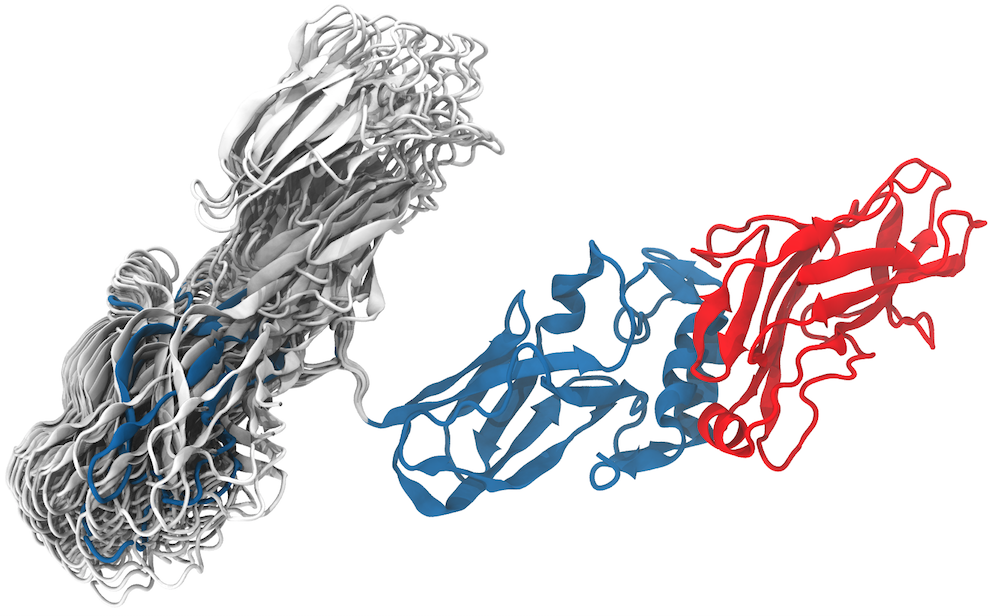
Fig. 4 - Generalized simulated annealing study of a cellulosome fragment. C. thermocellum CipA scaffolding fragment in complex with the SdbA type II cohesin module (CohII). CipA fragment components: ninth type I cohesin (CohI9), small flexible linker, X-module (Xmod) and type II dockerin (DocII). The analysis shows that the CohI9 module visited mainly two opposite positions. The positioning show how much the module can fluctuate around each conformation. In blue, one can observe the “native” position of the CohI9 module and the linker in the final minimized and equilibrated structure, initially taken from the crystal structure deposited in the PDB. The structure in blue was used to initiate all simulations and is not part of the results, it is, however, in the center of the region most visited by the independent simulations. In gray, a representation of significative structures showing the positions of final conformations from all 51,200 simulations. Read more |
Simulations reveal a tolerance to the positioning of the module in both conformations, which allows the cellulosome modules to adjust the large enzymatic domains attached to the type I cohesin. The results show that, even though very flexible, the linker led the type I Coh module to two preferred positions, with about 2/3 occurrences of the "native" conformation, as seen in the crystal structure, and 1/3 occurrences of the "alternate" conformation. The short flexible linker, one of the shortest in the C. thermocellum cellulosome, appears to have the function of keeping the system in two major conformations and not completely random ones as one might have expected. Read more on Biochimica et Biophysica Acta .

|
|
|
Fig. 5 - Illustration of celllosome environment. Animation adapted from Sasha Dale work. |
Ultrastable Interactions to Form a Large Enzymatic Machine
Challenging environments have guided nature in the development of ultrastable protein complexes, and cellulosomes are usually found in inhospitable circumstances. While network assembly of these large enzymatic complexes is enabled by protein interactions with commonplace affinities, it was found that certain cellulosomal ligand-receptor interactions exhibit extreme resistance to applied force (read more).
Employing state-of-the-art single-molecule atomic force microscope techniques and steered molecular dynamics simulations our group was able to characterize the ligand-receptor complex responsible for substrate anchoring in the Ruminococcus flavefaciens cellulosome, see Figure 5. The complex withstands forces of 600-750 pN, making it one of the strongest bimolecular interactions reported, equivalent to half the mechanical strength of a covalent bond.
Our findings, recently published on Nature Communications , demonstrate force activation and inter-domain stabilization of the complex, and suggest that certain network components serve as mechanical effectors for maintaining network integrity. The detailed understanding of cellulosomal network components may help in the development of biocatalysts for production of fuels and chemicals from renewable plant-derived biomass.
Molecular Mechanism of Cellulosomal Mechano-Stability
To elucidate the molecular mechanisms at play that enable the extreme mechanostability, steered molecular dynamics (SMD) simulations can be employed. The N-terminus of XMod-Doc was harmonically restrained while the C-terminus of Coh was pulled away at constant speed. SMD results showed the force increased with distance until the complex ruptured for all simulations. At the slowest pulling speed of 0.25 Å/ns the rupture occurred at a peak force of approximately 900 pN.
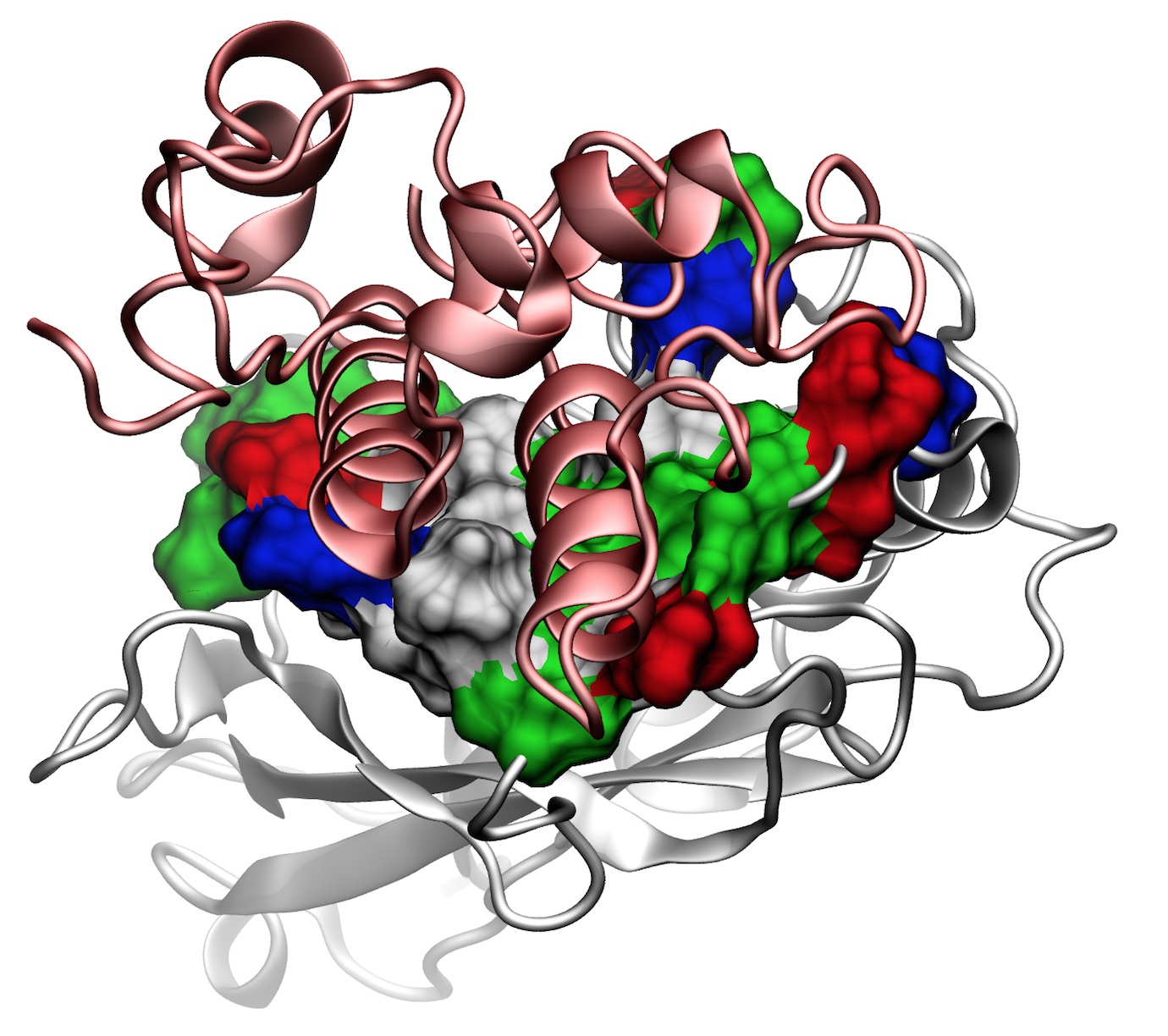
Molecular dynamics identified that the binding interface (see Figure 6) consisting of a hydrophobic centre (grey) surrounded by a ring of polar (green) and charged residues (blue, positive; red, negative). This residue pattern suggests the hydrophilic side chains protect the interior hydrophobic core from attack by water molecules, compensating for the flat binding interface that lacks a deep pocket. The geometry suggests a penalty to unbinding that stabilizes the bound state.
|
|
|
Fig. 6 - Illustration of the Cohesin-Dockerin binding interface. Secondary structure represetation in red shows the Doc domain over the Coh domain in grey. Surface representation in the Coh domain shows the amino-acids of the interface; hydrophobic residues (grey) surrounded by a ring of polar (green) and charged residues (blue, positive; red, negative). Read more |
|
|
|
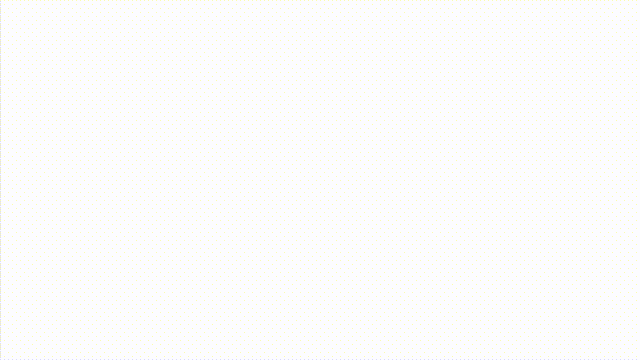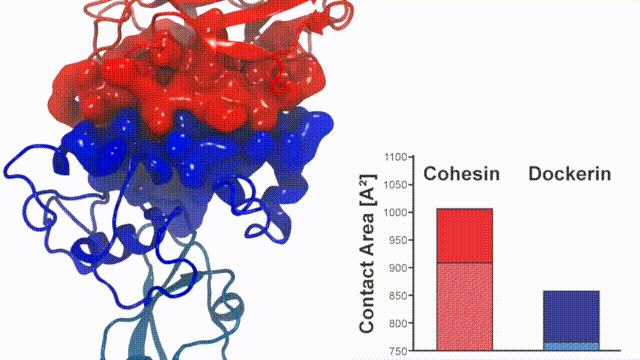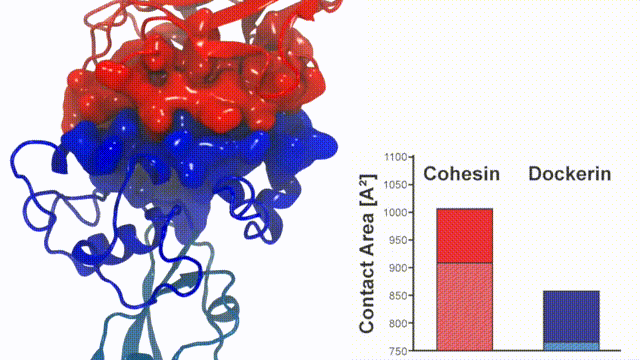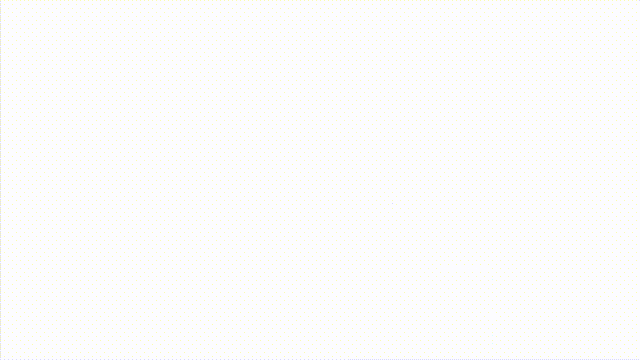
Fig. 7 - Illustration of the Cohesin-Dockerin contact. Cohesin domain in red; Dockerin and X-Module domains in blue. Under strain the adhesive bonds between cellulosome modules become stronger than seen in any other biomolecular system, in fact, become nearly as tight as strong chemical bonds. While the experimental data revealed bond strength and other characteristics, simulations reproducing the observed data provided a detailed view of the adhesive bond at atomic resolution, thereby revealing the physical mechanism underlying the uniquely adhesive property of cellulosomes. Read more |
Analysis of the contact surface areas of interacting residues shows that total contact area increase throughout the simulation due to rearrangement of the interacting residues when the complex is mechanically stressed,see Figure 7. Doc residues in the simulated binding interface clamped down on Coh residues upon mechanical loading, resulting in increased stability and decreased accessibility of water into the hydrophobic core of the bound complex.
Our results suggest that a catch bond mechanism is responsible for the remarkable stability under force and provide a molecular mechanism which the XMod-Doc:Coh complex uses to summon mechanical strength when needed, while still allowing relatively fast assembly and disassembly of the complex at equilibrium. Read more on Nature Communications
.Related publications
Ultrastable cellulosome-adhesion complex tightens under load
Constantin Schoeler, Klara H. Malinowska, Rafael C. Bernardi, Lukas F. Milles, Markus A. Jobst, Ellis Durner, Wolfgang Ott, Daniel B. Fried, Edward A. Bayer, Klaus Schulten, Hermann E. Gaub, and Michael A. Nash
Nature Communications 5:5635 (2014)
Enhanced sampling techniques in molecular dynamics simulations of biological systems
Rafael C. Bernardi, Marcelo C. R. Melo, and Klaus Schulten.
Biochimica et Biophysica Acta (2014)
Molecular dynamics study of enhanced Man5B enzymatic activity
Rafael C. Bernardi, Isaac Cann, and Klaus Schulten.
Biotechnology for Biofuels 7:83 (2014)
Investigators
Collaborators
Ed Bayer - Weizmann Institute of Science - Israel
Isaac Cann - Energy Biosciences Institute - University of Illinois at Urbana-Champaign
Douglas Clark - Energy Biosciences Institute - University of California - Berkeley
Matthew Francis - Energy Biosciences Institute - University of California - Berkeley
Hermann Gaub - Ludwig-Maximilians-Universitat Munchen - Germany
Support
Page created and maintained by Rafael Bernardi, Jodi Hadden and Joao Ribeiro.



