John E. Stone
- Distinguished Engineer, Scientific Visualization DevTech, NVIDIA
- Research Affiliate, Theoretical and Computational Biophysics Group and NIH Resource for Macromolecular Modeling and Visualization
Mr. Stone is an expert in scientific visualization, high performance ray tracing, and GPU computing techniques. Mr. Stone has served as the lead developer of VMD, a high performance tool for preparation, analysis, and visualization of biomolecular simulations used by over 100,000 researchers all over the world. Research papers describing VMD and its algorithms have been cited by more than 40,000 peer-reviewed research papers, both by users of VMD itself and by researchers developing related software and algorithms. Mr. Stone's research interests include molecular visualization, GPU computing, parallel computing, ray tracing, haptics, virtual environments, and immersive visualization. Mr. Stone is a frequent presenter and mentor in HPC and exascale computing training workshops and related "Hackathon" events that aim to give computational scientists both the knowledge of theory and the practical experience they need to successfully develop high performance scientific software for next-generation parallel computing systems. Mr. Stone was inducted as an NVIDIA CUDA Fellow in 2010. In 2015 Mr. Stone joined the Khronos Group Advisory Panel for the Vulkan Graphics API. In 2017, 2018, and 2019 Mr. Stone was awarded as an IBM Champion for Power, for innovative thought leadership in the technical community. In 2020, Mr. Stone joined the Khronos working group for ANARI, a new analytic rendering standard for high-fidelity scientific and technical visualization. Mr. Stone and coauthors won the ACM Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research at Supercomputing 2020. In 2021, Mr. Stone was a member of two finalist teams for the ACM Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research at Supercomputing 2021.
|
Google Scholar
ORCID Research Interests Research Articles Movies and Visualizations Book Chapters Presentations Service Press Articles Education Consulting Honors Linked-In Personal Site |

|
|

|
|
|
Contact:
Ph: 217-244-3349 johns@ks.uiuc.edu |
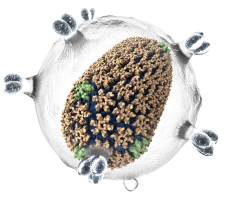
Over 1 billion-atom aerosolized
SARS-CoV-2 virion rendered with the VMD VdW and QuickSurf representations and the Tachyon parallel ray tracing engine |
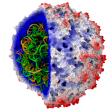
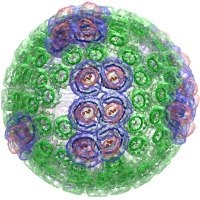
All-atom HIV-1 capsid
rendered with the VMD QuickSurf representation and the Tachyon parallel ray tracing engine |
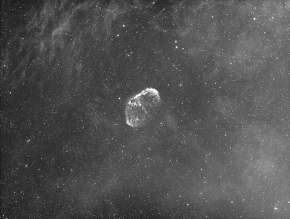
NGC6888 Crescent Nebula in Cygnus
313 min H-α exposure: 209x90sec subexposures Takahashi FSQ106N refractor, QHY163M camera, Astro-Physics AP1200GTO equatorial mount Urbana, IL, July 29, 2017 |
Research Interests:
VMD:
High performance visualization of biomolecular complexes,
interactive molecular dynamics simulations,
programmable shading
GPU Computing:
Accelerating scientific applications using graphics processing units
Tachyon:
Parallel ray tracing on GPUs, shared memory and
distributed memory architectures, molecular visualization
Immersive Visualization, Virtual Environments, and Haptic Feedback: CAVE, FreeVR, VRPN
Published Movies and Visualizations:
 |
|


|
|
 |
|
 |
|
 |
|
Published Journal and Book Covers, Visualizations, Photographs:










Book Chapters:
 |
|
 |
|
 |
|
 |
|
 |
|

|
|
 |
|
 |
|
 |
|
Research Articles:
- "VMD as a Platform for Interactive Small Molecule Preparation and Visualization for Quantum and Classical Simulations"
- Mariano Spivak, John E. Stone, João Ribeiro, Jan Saam, Peter L. Freddolino, Rafael. C. Bernardi, and Emad Tajkhorshid
- J. Chemical Information and Modeling, 63(15): 4664-4678, 2023.
- Online full text: https://doi.org/10.1021/acs.jcim.3c00658
- J. Chemical Information and Modeling, 63(15): 4664-4678, 2023.
- "Dynamics of Chromosome Organization in a Minimal Bacterial Cell"
- Benjamin R. Gilbert, Zane R. Thornburg, Troy A. Brier, Jan A. Stevens, Fabian Grunewald, John E. Stone, Siewert Marrink, Zaida A. Luthey-Schulten
- Frontiers in Cell and Developmental Biology 11, 1214962, 2023.
- Online full text: https://doi.org/10.3389/fcell.2023.1214962
- Frontiers in Cell and Developmental Biology 11, 1214962, 2023.
- "Application Experiences on a GPU-Accelerated Arm-based HPC Testbed"
- Wael Elwasif, William Godoy, Nick Hagerty, J Austin Harris, Oscar Hernandez, Balint Joo, Paul Kent, Damien Lebrun-Grandie, Elijah Maccarthy, Veronica Melesse Vergara, Bronson Messer, Ross Miller, Sarp Oral, Sergei Bastrakov, Michael Bussmann, Alexander Debus, Klaus Steiniger, Jan Stephan, Rene Widera, Spencer Bryngelson, Henry Le Berre, Anand Radhakrishnan, Jeffrey Young, Sunita Chandrasekaran, Florina Ciorba, Osman Simsek, Kate Clark, Filippo Spiga, Jeff Hammond, John Stone, David Hardy, Sebastian Keller, Jean-Guillaume Piccinali, Christian Trott
- HPC Asia '23 Workshops: Proceedings of the HPC Asia 2023 Workshops, pp. 35-49, 2023.
- Online full text: https://doi.org/10.1145/3581576.3581621
- HPC Asia '23 Workshops: Proceedings of the HPC Asia 2023 Workshops, pp. 35-49, 2023.
- "Early Application Experiences on GPU-Accelerated ARM HPC Platforms"
- Wael Elwasif, William Godoy, Nick Hagerty, J Austin Harris, Oscar Hernandez, Balint Joo, Paul Kent, Damien Lebrun-Grandie, Elijah Maccarthy, Veronica Melesse Vergara, Bronson Messer, Ross Miller, Sarp Oral, Sergei Bastrakov, Michael Bussmann, Alexander Debus, Klaus Steiniger, Jan Stephan, Rene Widera, Spencer Bryngelson, Henry Le Berre, Anand Radhakrishnan, Jeffrey Young, Sunita Chandrasekaran, Florina Ciorba, Osman Simsek, Kate Clark, Filippo Spiga, Jeff Hammond, John Stone, David Hardy, Sebastian Keller, Jean-Guillaume Piccinali, Christian Trott
- arXiv:2209.09731, Dec 19, 2022.
- Online full text: https://doi.org/10.48550/arXiv.2209.09731
- arXiv:2209.09731, Dec 19, 2022.
- "py-MCMD: Python Software for Performing Hybrid Monte Carlo -- Molecular Dynamics Simulations with GOMC and NAMD"
- Mohammad Soroush Barhaghi, Brad Crawford, Gregory Schwing, David J. Hardy, John E. Stone, Loren Schwiebert, Jeffrey Potoff, Emad Tajkhorshid.
- Journal of Chemical Theory and Computation, 18: 4983-4994, 2022.
- Online full text: https://doi.org/10.1021/acs.jctc.1c00911
- Journal of Chemical Theory and Computation, 18: 4983-4994, 2022.
- "Analytic Rendering and Hardware-Accelerated Simulation for Scientific Applications"
- Paul Navratil, Christiaan Gribble, Pascal Grossett, John E. Stone.
- Computing in Science and Engineering, 24(2):4-6, 2022.
- Online full text: https://doi.org/10.1109/MCSE.2022.3163480
- Computing in Science and Engineering, 24(2):4-6, 2022.
- "ANARI: A 3D Rendering API Standard"
- John E. Stone, Kevin Griffin, Jefferson Amstutz, David E. DeMarle, William Sherman, Johannes Günther.
- Computing in Science and Engineering, 24(2):7-18, 2022.
- Online full text: https://doi.org/10.1109/MCSE.2022.3163151
- Computing in Science and Engineering, 24(2):7-18, 2022.
- "Intelligent Resolution: Integrating Cryo-EM with AI-Driven Multi-Resolution Simulations to Observe the SARS-CoV-2 Replication-Transcription Machinery in Action"
- Andra Trifan, Defne Gorgun, Zongyi Li, Alexander Brace, Maxim Zvyagin,
Heng Ma, Austin Clyde, David Clark, Michael Salim, David J. Hardy,
Tom Burnley, Lei Huang, John McCalpin, Murali Emani, Hyenseung Yoo,
Junqi Yin, Aristeidis Tsaris, Vishal Subbiah, Tanveer Raza, Jessica Liu,
Noah Trebesch, Geoffrey Wells, Venkatesh Mysore, Thomas Gibbs,
James Phillips, S. Chakra Chennubhotla, Ian Foster, Rick Stevens,
Anima Anandkumar, Venkatram Vishwanath, John E. Stone,
Emad Tajkhorshid, Sarah A. Harris, Arvind Ramanathan.
- International Journal of High Performance Computing Applications (IJHPCA), 36(5-6):603-623, 2022.
- Finalist, ACM Gordon Bell Special Prize for COVID-19 Research, 2021.
- Overview of our Gordon Bell team's research
- Online full text https://doi.org/10.1177/10943420221113513
- Online full text pre-print available at bioRxiv: https://www.biorxiv.org/content/10.1101/2021.10.09.463779v1
- International Journal of High Performance Computing Applications (IJHPCA), 36(5-6):603-623, 2022.
- "#COVIDisAirborne: AI-Enabled Multiscale Computational Microscopy of Delta SARS-CoV-2 in a Respiratory Aerosol"
- Abigail Dommer, Lorenzo Casalino, Fiona Kearns, Mia Rosenfeld,
Nicholas Wauer, Surl-Hee Ahn, John Russo, Sofia Oliveira,
Clare Morris, Anthony Bogetti, Anda Trifan, Alexander Brace,
Terra Sztain, Austin Clyde, Heng Ma, Chakra Chennubhotla,
Hyungro Less, Matteo Turilli, Syma Khalid, Terasa Tamayo-Mendoza,
Matthew Welborn, Anders Christensen, Daniel G. A. Smith,
Zhuoran Qiao, Sai Krishna Sirumalla, Michael O'Connor,
Frederick Manby, Anima Anandkumar, David J. Hardy, James C. Phillips,
Abraham Stern, Josh Romero, David Clark, Mitchell Dorrell, Tom Maiden,
Lei Huang, John McCalpin, Christopher Woods, Matt Williams,
Bryan Barker, Harinda Rajapaksha, Richard Pitts, Tom Gibbs,
John E. Stone, Daniel Zuckerman, Adrian Mulholland, Thomas Miller III,
Shantenu Jha, Arvind Ramanathan, Lillian Chong, Rommie Amaro.
- International Journal of High Performance Computing Applications (IJHPCA), 2022. (To appear)
- Finalist, ACM Gordon Bell Special Prize for COVID-19 Research, 2021.
- Overview of our Gordon Bell team's research
- Online full text https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8609898/
- Online full text pre-print available at bioRxiv: https://www.biorxiv.org/content/10.1101/2021.11.12.468428v1
- International Journal of High Performance Computing Applications (IJHPCA), 2022. (To appear)
- "Lessons Learned from Responsive Molecular Dynamics Studies of the COVID-19 Virus"
- David J. Hardy, John E. Stone, Barry Isralewitz, Emad Tajkhorshid
- 2021 IEEE/ACM HPC for Urgent Decision Making (UrgentHPC), pp. 1-10, 2021.
- Online full text https://dx.doi.org/10.1109/UrgentHPC54802.2021.00006
- 2021 IEEE/ACM HPC for Urgent Decision Making (UrgentHPC), pp. 1-10, 2021.
- "AI-Driven Multiscale Simulations Illuminate Mechanisms of SARS-CoV-2 Spike Dynamics"
- Lorenzo Casalino, Abigail Dommer, Zied Gaieb, Emilia P. Barros,
Terra Sztain, Surl-Hee Ahn, Anda Trifan, Alexander Brace,
Anthony Bogetti, Heng Ma, Hyungro Lee, Matteo Turilli, Syma Khalid,
Lillian Chong, Carlos Simmerling, David J. Hardy, Julio D. C. Maia,
James C. Phillips, Thorsten Kurth, Abraham Stern, Lei Huang,
John McCalpin, Mahidhar Tatineni, Tom Gibbs, John E. Stone,
Shantenu Jha, Arvind Ramanathan, Rommie E. Amaro.
- International Journal of High Performance Computing Applications (IJHPCA), 35(5), April 2021.
- Winner of the ACM Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research
- Overview of our Gordon Bell team's research
- Online full text https://dx.doi.org/10.1177/10943420211006452
- Online full text pre-print available at bioRxiv: https://www.biorxiv.org/content/10.1101/2020.11.19.390187v1
- International Journal of High Performance Computing Applications (IJHPCA), 35(5), April 2021.
- "Multiscale modeling and cinematic visualization of photosynthetic energy conversion processes from electronic to cell scales"
- Melih Sener, Stuart Levy, John E. Stone, AJ Christensen,
Barry Isralewitz, Robert Patterson, Kalina Borkiewicz,
Jeffrey Carpenter, C. Neil Hunter, Zaida Luthey-Schulten, Donna Cox
- J. Parallel Computing, 102, pp. 102698, 2021.
- Online full text available at https://doi.org/10.1016/j.parco.2020.102698
- J. Parallel Computing, 102, pp. 102698, 2021.
- "Scalable molecular dynamics on CPU and GPU architectures with NAMD"
- James C. Phillips, David J. Hardy, Julio D. C. Maia, John E. Stone,
João V. Ribeiro, Rafael C. Bernardi, Ronak Buch, Giacomo Fiorin,
Jérôme Hénin, Wei Jiang, Ryan McGreevy, Marcelo C. R. Melo,
Brian K. Radak, Robert D. Skeel, Abhishek Singharoy, Yi Wang,
Benoit Roux, Aleksei Aksimentiev, Zaida Luthey-Schulten,
Laxmikant V. Kalé, Klaus Schulten, Christophe Chipot, Emad Tajkhorshid.
- J. Chemical Physics, 153(4), 2020.
- Online full text available at https://dx.doi.org/10.1063/5.0014475
- J. Chemical Physics, 153(4), 2020.
- "Scalable Analysis of Authentic Viral Envelopes on FRONTERA"
- Fabio Gonzalez-Arias, Tyler Reddy, John E. Stone, Jodi A. Hadden-Perilla, and Juan R. Perilla.
- Computing in Science and Engineering, 22(6):11-20, 2020. Winner, CiSE 2020 Best Paper Award
- Online full text available at https://dx.doi.org/10.1109/MCSE.2020.3020508
- Computing in Science and Engineering, 22(6):11-20, 2020. Winner, CiSE 2020 Best Paper Award
- "An accessible visual narrative for the primary energy source of life from the fulldome show Birth of Planet Earth"
- Melih Sener, Stuart Levy, AJ Chistensen, Robert Patterson,
Kalina Borkiewicz, John E. Stone, Barry Isralewitz,
Jeffrey Carpenter, Donna Cox.
- SC'19 Visualization and Data Analytics Showcase, 2019.
- Winner of the SC'19 Visualization and Data Analytics Showcase
- SC'19 Visualization and Data Analytics Showcase
- SC'19 Visualization and Data Analytics Showcase, 2019.
- "Atoms to Phenotypes: Molecular Design Principles of Cellular Energy Metabolism"
- A. Singharoy, C. Maffeo, K. H. Delgado-Magnero, D. J. K. Swainsbury, M. Sener, U. Kleinekathöfer, B. Isralewitz, I. Teo, D. Chandler, J. E. Stone, J. Phillips, T. V. Pogorelov, M. I. Mallus, C. Chipot, Z. Luthey-Schulten, P. Tieleman, C. N. Hunter, E. Tajkhorshid, A. Aksimentiev, K. Schulten.
- Cell 179(5), pp. 1098-1111.e23, November 14, 2019.
- Online full text available at https://doi.org/10.1016/j.cell.2019.10.021
- Cell 179(5), pp. 1098-1111.e23, November 14, 2019.
- "High-Performance Analysis of Biomolecular Containers to Measure Small-Molecule Transport, Transbilayer Lipid Diffusion, and Protein Cavities"
- Alexander J. Bryer, Jodi A. Hadden, John E. Stone, Juan R. Perilla.
- J. Chemical Information and Modeling, 59(10), 4328-4338, 2019.
- Online full text available at http://dx.doi.org/10.1021/acs.jcim.9b00324
- J. Chemical Information and Modeling, 59(10), 4328-4338, 2019.
- "NanoShaper-VMD interface: computing and visualizing surfaces, pockets and channels in molecular systems"
- Sergio Decherchi, Andrea Spitaleri, John Stone, and Walter Rocchia.
- Bioinformatics 35(7): 1241-1243, 2019.
- Online full text available at http://dx.doi.org/10.1093/bioinformatics/bty761
- Bioinformatics 35(7): 1241-1243, 2019.
- "Scalable Molecular Dynamics with NAMD on the Summit System"
- Bilge Acun, David J. Hardy, Laxmikant V. Kale, Ke Li, James C. Phillips, and John E. Stone.
- IBM Journal of Research and Development, 62(6):4:1-4:9, 2018.
- Online full text available at http://dx.doi.org/10.1147/JRD.2018.2888986
- IBM Journal of Research and Development, 62(6):4:1-4:9, 2018.
- "Best Practices in Running Collaborative GPU Hackathons: Advancing Scientific Applications with a Sustained Impact"
- Sunita Chandrasekaran, Guido Juckeland, Meifeng Ling, Matthew Otten, Dirk Pleiter, John E. Stone, Juan Lucio-Vega, Michael Zingale, and Fernanda Foertter.
- Computing in Science and Engineering 20(4):95-106, 2018.
- Online full text available at http://dx.doi.org/10.1109/MCSE.2018.042781332
- Computing in Science and Engineering 20(4):95-106, 2018.
- "NAMD goes quantum: an integrative suite for hybrid simulations"
- Marcelo C. R. Melo, Rafael C. Bernardi, Till Rudack, Maximilian Scheurer, Christoph Riplinger, James C. Phillips, Julio D. C. Maia, Gerd B. Rocha, João V. Ribeiro, John E. Stone, Frank Neese, Klaus Schulten, and Zaida Luthey-Schulten.
- Nature Methods, 15: 351-354, 2018.
- Online full text available at https://www.nature.com/articles/nmeth.4638
- Nature Methods, 15: 351-354, 2018.
- "The OLCF GPU Hackathon Series: The Story Behind Advancing Scientific Applications with a Sustained Impact"
- Sunita Chandrasekaran, Guido Juckeland, Meifeng Ling, Matthew Otten, Dirk Pleiter, John E. Stone, Juan Lucio-Vega, Michael Zingale, and Fernanda Foertter.
- The 2017 Workshop on Education for High Performance Computing (EduHPC'17), 2017.
- The 2017 Workshop on Education for High Performance Computing (EduHPC'17), 2017.
- "Challenges of Integrating Stochastic Dynamics and Cryo-electron Tomograms in Whole-cell Simulations"
- Tyler M. Earnest, Reika Watanabe, John E. Stone, Julia Mahamid, Wolfgang Baumeister, Elizabeth Villa, and Zaida Luthey-Schulten.
- J. Physical Chemistry B, 121(15): 3871-3881, 2017.
- Online full text available at http://pubs.acs.org/doi/10.1021/acs.jpcb.7b00672
- J. Physical Chemistry B, 121(15): 3871-3881, 2017.
- "Early Experiences Porting the NAMD and VMD Molecular Simulation and Analysis Software to GPU-Accelerated OpenPOWER Platforms"
- John E. Stone, Antti-Pekka Hynninen, James C. Phillips, and Klaus Schulten.
- International Workshop on OpenPOWER for HPC (IWOPH'16), LNCS 9945, pp. 188-206, 2016.
- Online full text available at http://dx.doi.org/10.1007/978-3-319-46079-6_14
- International Workshop on OpenPOWER for HPC (IWOPH'16), LNCS 9945, pp. 188-206, 2016.
-
"Molecular dynamics-based refinement and validation for sub-5Å cryo-electron microscopy maps" - Abhishek Singharoy, Ivan Teo, Ryan McGreevy, John E. Stone, Jianhua Zhao, and Klaus Schulten.
- eLife 2016;10.7554/eLife.16105.
- Online full text available at http://dx.doi.org/10.7554/eLife.16105

- Abhishek Singharoy, Ivan Teo, Ryan McGreevy, John E. Stone, Jianhua Zhao, and Klaus Schulten.
- "TopoGromacs: Automated Topology Conversion from CHARMM to GROMACS within VMD"
- Josh V. Vermaas, David J. Hardy, John E. Stone, Emad Tajkhorshid, and Axel Kohlmeyer.
- J. Chemical Information and Modeling, 56(6):1112-1116, 2016.
- Online full text available at http://dx.doi.org/10.1021/acs.jcim.6b00103
- J. Chemical Information and Modeling, 56(6):1112-1116, 2016.
- "QwikMD - Integrative Molecular Dynamics Toolkit for Novices and Experts"
- João V. Ribeiro, Rafael C. Bernardi, Till Rudack, John E. Stone, James C. Phillips, Peter L. Freddolino, and Klaus Schulten.
- Scientific Reports, 6:2653, 2016.
- Online full text available at http://dx.doi.org/10.1038/srep26536
- Scientific Reports, 6:2653, 2016.
- "Immersive Molecular Visualization with Omnidirectional Stereoscopic Ray Tracing and Remote Rendering"
- John E. Stone, William R. Sherman, and Klaus Schulten.
- High Performance Data Analysis and Visualization Workshop, 2016 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW), pp. 1048-1057, 2016.
- Online full text available at http://dx.doi.org/10.1109/IPDPSW.2016.121
- High Performance Data Analysis and Visualization Workshop, 2016 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW), pp. 1048-1057, 2016.
- "High Performance Molecular Visualization: In-Situ and Parallel Rendering with EGL"
- John E. Stone, Peter Messmer, Robert Sisneros, and Klaus Schulten.
- High Performance Data Analysis and Visualization Workshop, 2016 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW), pp. 1014-1023, 2016.
- Online full text available at http://dx.doi.org/10.1109/IPDPSW.2016.127
- High Performance Data Analysis and Visualization Workshop, 2016 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW), pp. 1014-1023, 2016.
- "Evaluation of Emerging Energy-Efficient Heterogeneous Computing Platforms for Biomolecular and Cellular Simulation Workloads"
- John E. Stone, Michael J. Hallock, James C. Phillips,
Joseph R. Peterson, Zaida Luthey-Schulten, and Klaus Schulten.
- 25th International Heterogeneity in Computing Workshop, 2016 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW), pp. 89-100, 2016.
- Online full text available at http://dx.doi.org/10.1109/IPDPSW.2016.130
- 25th International Heterogeneity in Computing Workshop, 2016 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW), pp. 89-100, 2016.
- "Atomic Detail Visualization of Photosynthetic Membranes with GPU-Accelerated Ray Tracing"
- John E. Stone, Melih Sener, Kirby L. Vandivort, Angela Barragan,
Abhishek Singharoy, Ivan Teo, João V. Ribeiro, Barry Isralewitz,
Bo Liu, Boon Chong Goh, James C. Phillips, Craig MacGregor-Chatwin,
Matthew P. Johnson, Lena F. Kourkoutis, C. Neil Hunter, and Klaus Schulten
- J. Parallel Computing, 55:17-27, 2016.
- Online full text available at http://dx.doi.org/10.1016/j.parco.2015.10.015
- J. Parallel Computing, 55:17-27, 2016.
- "Chemical Visualization of Human Pathogens: the Retroviral Capsids"
- Juan R. Perilla, Boon Chong Goh, John E. Stone, and Klaus Schulten
- SC'15 Visualization and Data Analytics Showcase, 2015.
- Finalist, SC'15 Visualization and Data Analytics Showcase
- Online full text available at SC'15 site
- SC'15 Visualization and Data Analytics Showcase, 2015.
- "Runtime and Architecture Support for Efficient Data Exchange in Multi-Accelerator Applications"
- Javier Cabezas, Isaac Gelado, John E. Stone,
Nacho Navarro, David B. Kirk, and Wen-mei Hwu.
- IEEE Transactions on Parallel and Distributed Systems, 26(5):1405-1418, 2015.
- Online full text available at http://dx.doi.org/10.1109/TPDS.2014.2316825
- IEEE Transactions on Parallel and Distributed Systems, 26(5):1405-1418, 2015.
- "Multilevel Summation Method for Electrostatic Force Evaluation"
- David J. Hardy, Zhe Wu, James C. Phillips John E. Stone,
Robert D. Skeel, and Klaus Schulten.
- Journal of Chemical Theory and Computation, 11:766-779, 2015.
- Online full text available at http://pubs.acs.org/doi/abs/10.1021/ct5009075
- Journal of Chemical Theory and Computation, 11:766-779, 2015.
- "Visualization of Energy Conversion Processes in a Light Harvesting Organelle at Atomic Detail"
- Melih Sener, John E. Stone, Angela Barragan, Abhishek Singharoy,
Ivan Teo, Kirby L. Vandivort, Barry Isralewitz, Bo Liu,
Boon Chong Goh, James C. Phillips,
Lena F. Kourkoutis, C. Neil Hunter, and Klaus Schulten
- SC'14 Visualization and Data Analytics Showcase, 2014.
- Winner of the SC'14 Visualization and Data Analytics Showcase
- Online full text available at SC'14 site
- SC'14 Visualization and Data Analytics Showcase, 2014.
- "Petascale Tcl with NAMD, VMD, and Swift/T"
- James C. Phillips, John E. Stone, Kirby L. Vandivort,
Timothy G. Armstrong, Justin M. Wozniak, Michael Wilde,
and Klaus Schulten.
- Proceedings of the First Workshop for High Performance Technical Computing in Dynamic Languages, pp. 6-17, 2014.
- Online full text available at ACM digital library
- Online full text available at http://dx.doi.org/10.1109/HPTCDL.2014.7
- Proceedings of the First Workshop for High Performance Technical Computing in Dynamic Languages, pp. 6-17, 2014.
- "Stable Small Quantum Dots for Synaptic Receptor Tracking on Live Neurons"
- En Cai, Pinghua Ge, Sang Hak Lee, Okunola Jeyifous, Yong Wang,
Yanxin Liu, Katie M. Wilson, Sung Jun Lim, Michele A. Baird,
John E. Stone, Kwan Young Lee, David G. Fernig, Michael W. Davidson,
Hee Jung Chung, Klaus Schulten, Andrew M. Smith, William N. Green,
and Paul R. Selvin.
- Angewandte Chemie - International Edition in English, 53(46):12484-12488, 2014.
- Online full text available at http://dx.doi.org/10.1002/anie.201405735
- Angewandte Chemie - International Edition in English, 53(46):12484-12488, 2014.
- "Unlocking the Full Potential of the Cray XK7 Accelerator"
- Mark D. Klein and John E. Stone.
- Cray Users Group, Lugano Switzerland, May 2014.
- Online full text in CUG 2014 Proceedings
- Cray Users Group, Lugano Switzerland, May 2014.
- "GPU-Accelerated Analysis and Visualization of Large Structures Solved by Molecular Dynamics Flexible Fitting"
- John E. Stone, Ryan McGreevy, Barry Isralewitz, and Klaus Schulten.
- Faraday Discussions, 169:265-283, 2014.
- Online full text available at http://dx.doi.org/10.1039/C4FD00005F
- Faraday Discussions, 169:265-283, 2014.
- "Methodologies for the Analysis of Instantaneous Lipid Diffusion in MD Simulations of Large Membrane Systems"
- Matthieu Chavent, Tyler Reddy, Joseph Goose, Anna Caroline E. Dahl,
John E. Stone, Bruno Jobard, and Mark S.P. Sansom.
- Faraday Discussions, 169:455-475, 2014.
- Online full text available at http://dx.doi.org/10.1039/C3FD00145H
- Faraday Discussions, 169:455-475, 2014.
- "Simulation of reaction diffusion processes over biologically relevant size and time scales using multi-GPU workstations"
- Michael J. Hallock, John E. Stone, Elijah Roberts,
Corey Fry, and Zaida Luthey-Schulten.
- Journal of Parallel Computing, 40:86-99, 2014.
- Online full text available at http://dx.doi.org/10.1016/j.parco.2014.03.009
- Journal of Parallel Computing, 40:86-99, 2014.
- "Assembly of Nsp1 nucleoporins provides insight into nuclear pore complex gating"
- Ramya Gamini, Wei Han, John E. Stone, and Klaus Schulten.
- PLoS Computational Biology, 10(3): e1003488, 2014.
- Online full text available at http://dx.doi.org/10.1371/journal.pcbi.1003488
- PLoS Computational Biology, 10(3): e1003488, 2014.
- "GPU-Accelerated Molecular Visualization on Petascale Supercomputing Platforms"
- John E. Stone, Kirby L. Vandivort, and Klaus Schulten.
- UltraVis'13: Proceedings of the 8th International Workshop on Ultrascale Visualization, pp. 6:1-6:8, 2013.
- Online full text available at http://dx.doi.org/10.1145/2535571.2535595
- UltraVis'13: Proceedings of the 8th International Workshop on Ultrascale Visualization, pp. 6:1-6:8, 2013.
- "Early Experiences Scaling VMD Molecular Visualization and Analysis Jobs on Blue Waters"
- John E. Stone, Barry Isralewitz, and Klaus Schulten.
- Extreme Scaling Workshop (XSW), pp. 43-50, 2013.
- Online full text available at http://dx.doi.org/10.1109/XSW.2013.10
- Online full text available at http://www.xsede.org/web/xscale/agenda
- Extreme Scaling Workshop (XSW), pp. 43-50, 2013.
- "Lattice microbes: High-Performance stochastic simulation method for the reaction-diffusion master equation"
- Elijah Roberts, John E. Stone, and Zaida Luthey-Schulten.
- Journal of Computational Chemistry, 34(3):245-255, 2013.
- Online full text: http://dx.doi.org/10.1002/jcc.23130
- Journal of Computational Chemistry, 34(3):245-255, 2013.
- "Fast Visualization of Gaussian Density Surfaces for Molecular Dynamics and Particle System Trajectories"
- Michael Krone, John E. Stone, Thomas Ertl, and Klaus Schulten.
- EuroVis - Short Papers, pp. 67-71, 2012.
- Online full text: http://dx.doi.org/10.2312/PE/EuroVisShort/EuroVisShort2012/067-071
- EuroVis - Short Papers, pp. 67-71, 2012.
- "Optimization of a Broadband Discone Antenna Design and Platform Installed Radiation Patterns Using a GPU-Accelerated Savant/WIPL-D Hybrid Approach"
- Tod Courtney, Matthew C. Miller, John E. Stone, and Robert A. Kipp.
- In Proceedings of the Applied Computational Electromagnetics Symposium (ACES 2012), Columbus, Ohio, April 2012.
- Online full text
- In Proceedings of the Applied Computational Electromagnetics Symposium (ACES 2012), Columbus, Ohio, April 2012.
- "Immersive Out-of-Core Visualization of Large-Size and Long-Timescale Molecular Dynamics Trajectories"
- John E. Stone, Kirby L. Vandivort, and Klaus Schulten.
- G. Bebis et al. (Eds.): 7th International Symposium on Visual Computing (ISVC 2011), LNCS 6939, pp. 1-12, 2011.
- Online full text: http://dx.doi.org/10.1007/978-3-642-24031-7_1
- G. Bebis et al. (Eds.): 7th International Symposium on Visual Computing (ISVC 2011), LNCS 6939, pp. 1-12, 2011.
- "Using GPUs to Accelerate Installed Antenna Performance Simulations"
- Tod Courtney, John E. Stone, and Bob Kipp.
- In Proceedings of the 2011 Antenna Applications Symposium, Allerton Park, Monticello IL, September 2011.
- Online full text
- In Proceedings of the 2011 Antenna Applications Symposium, Allerton Park, Monticello IL, September 2011.
- "Fast Analysis of Molecular Dynamics Trajectories with Graphics Processing Units — Radial Distribution Function Histogramming"
- Benjamin G. Levine, John E. Stone, and Axel Kohlmeyer.
- Journal of Computational Physics, 230(9):3556-3569, 2011.
- Online full text: http://dx.doi.org/10.1016/j.jcp.2011.01.048
- Journal of Computational Physics, 230(9):3556-3569, 2011.
- "Immersive Molecular Visualization and Interactive Modeling with Commodity Hardware"
- John E. Stone, Axel Kohlmeyer, Kirby L. Vandivort, and Klaus Schulten.
- G. Bebis et al. (Eds.): ISVC 2010, Part II, LNCS 6454, pp. 382-393, 2010.
- Online full text: http://dx.doi.org/10.1007/978-3-642-17274-8_38
- G. Bebis et al. (Eds.): ISVC 2010, Part II, LNCS 6454, pp. 382-393, 2010.
- "Quantifying the Impact of GPUs on Performance and Energy Efficiency in HPC Clusters"
- Jeremy Enos, Craig Steffen, Joshi Fullop, Michael Showerman,
Guochun Shi, Kenneth Esler, Volodymyr Kindratenko,
John E. Stone, and James C. Phillips.
- International Conference on Green Computing, pp. 317-324, 2010.
- Online full text: http://dx.doi.org/10.1109/GREENCOMP.2010.5598297
- International Conference on Green Computing, pp. 317-324, 2010.
- "GPU-Accelerated Molecular Modeling Coming Of Age"
- John E. Stone, David J. Hardy, Ivan S. Ufimtsev, and Klaus Schulten.
- Journal of Molecular Graphics and Modelling, 29(2):116-125, 2010.
- Online full text: http://dx.doi.org/10.1016/j.jmgm.2010.06.010
- Journal of Molecular Graphics and Modelling, 29(2):116-125, 2010.
- "OpenCL: A Parallel Programming Standard for Heterogeneous Computing Systems"
- John E. Stone, David Gohara, and Guochun Shi.
- Computing in Science and Engineering, 12(3):66-73, 2010.
- Online full text: http://dx.doi.org/10.1109/MCSE.2010.69
- Computing in Science and Engineering, 12(3):66-73, 2010.
- "An Asymmetric Distributed Shared Memory Model for Heterogeneous Parallel Systems"
- Isaac Gelado, John E. Stone, Javier Cabezas, Sanjay Patel, Nacho Navarro, and Wen-mei W. Hwu.
- ASPLOS '10: Proceedings of the 15th International Conference on Architectural Support for Programming Languages and Operating Systems, pp. 347-358, 2010.
- Online full text: http://doi.acm.org/10.1145/1736020.1736059
- ASPLOS '10: Proceedings of the 15th International Conference on Architectural Support for Programming Languages and Operating Systems, pp. 347-358, 2010.
- "Probing Biomolecular Machines with Graphics Processors"
- James C. Phillips and John E. Stone.
- Communications of the ACM 52(10):34-41, 2009.
- Online full text: http://doi.acm.org/10.1145/1562764.1562780
- Communications of the ACM 52(10):34-41, 2009.
- "GPU Clusters for High Performance Computing"
- Volodymyr Kindratenko, Jeremy Enos, Guochun Shi, Michael Showerman,
Galen Arnold, John E. Stone, James Phillips, and Wen-mei Hwu.
- Cluster Computing and Workshops, 2009. CLUSTER '09. IEEE International Conference on. pp. 1-8, Aug. 2009.
- Online full text: http://dx.doi.org/10.1109/CLUSTR.2009.5289128
- Cluster Computing and Workshops, 2009. CLUSTER '09. IEEE International Conference on. pp. 1-8, Aug. 2009.
- "Visualization of Cyclic and Multi-branched Molecules with VMD"
- Simon Cross, Michelle M. Kuttell, John E. Stone, and James E. Gain.
- Journal of Molecular Graphics and Modelling. 28:131-139, 2009.
- Online full text: http://dx.doi.org/10.1016/j.jmgm.2009.04.010
- Journal of Molecular Graphics and Modelling. 28:131-139, 2009.
- "Long time-scale simulations of in vivo diffusion using GPU hardware"
- Elijah Roberts, John E. Stone, Leonardo Sepulveda, Wen-mei W. Hwu, and Zaida Luthey-Schulten.
- IPDPS '09: Proceedings of the 2009 IEEE International Symposium on Parallel & Distributed Processing, pp. 1-8, 2009
- Online full text: http://dx.doi.org/10.1109/IPDPS.2009.5160930
- IPDPS '09: Proceedings of the 2009 IEEE International Symposium on Parallel & Distributed Processing, pp. 1-8, 2009
- "High Performance Computation and Interactive Display of Molecular Orbitals on GPUs and Multi-core CPUs"
- John E. Stone, Jan Saam, David J. Hardy, Kirby L. Vandivort,
Wen-mei W. Hwu, and Klaus Schulten.
- In Proceedings of the 2nd Workshop on General-Purpose Processing on Graphics Processing Units, ACM International Conference Proceeding Series, volume 383, pp. 9-18, 2009.
- Online full text: http://doi.acm.org/10.1145/1513895.1513897
- In Proceedings of the 2nd Workshop on General-Purpose Processing on Graphics Processing Units, ACM International Conference Proceeding Series, volume 383, pp. 9-18, 2009.
- "Multilevel Summation of Electrostatic Potentials Using Graphics Processing Units"
- David J. Hardy, John E. Stone, and Klaus Schulten.
- Parallel Computing, 28:164-177, 2009.
- Online full text: http://dx.doi.org/10.1016/j.parco.2008.12.005
- Parallel Computing, 28:164-177, 2009.
- "Adapting a Message-driven Parallel Application to GPU-Accelerated Clusters"
- James C. Phillips, John E. Stone, and Klaus Schulten.
- In SC '08: Proceedings of the 2008 ACM/IEEE Conference on Supercomputing, pp. 1-9, Piscataway, NJ, USA, 2008. IEEE Press.
- Online full text: http://dx.doi.org/10.1109/SC.2008.5214716
- In SC '08: Proceedings of the 2008 ACM/IEEE Conference on Supercomputing, pp. 1-9, Piscataway, NJ, USA, 2008. IEEE Press.
- "Using VMD - An Introductory Tutorial"
- Jen Hsin, Anton Arkhipov, Ying Yin, John E. Stone, and Klaus Schulten.
- Current Protocols - Bioinformatics, 5:Unit 5.7, 2008
- Online full text: http://dx.doi.org/10.1002/0471250953.bi0507s24
- Current Protocols - Bioinformatics, 5:Unit 5.7, 2008
- "GPU Acceleration of Cutoff Pair Potentials for Molecular Modeling Applications"
- Christopher I. Rodrigues, David J. Hardy, John E. Stone,
Klaus Schulten, and Wen-Mei W. Hwu.
- In CF'08: Proceedings of the 2008 conference on Computing frontiers, pp. 273-282, New York, NY, USA, 2008. ACM.
- Online full text: http://doi.acm.org/10.1145/1366230.1366277
- In CF'08: Proceedings of the 2008 conference on Computing frontiers, pp. 273-282, New York, NY, USA, 2008. ACM.
- "GPU Computing"
- John D. Owens, Mike Houston, David Luebke, Simon Green, John E. Stone, and James C. Phillips.
- Proceedings of the IEEE, 96:879-899, 2008.
- Online full text: http://dx.doi.org/10.1109/JPROC.2008.917757
- Proceedings of the IEEE, 96:879-899, 2008.
- "Accelerating Molecular Modeling Applications with Graphics Processors"
- John E. Stone, James C. Phillips, Peter L. Freddolino, David J. Hardy, Leonardo G. Trabuco, and Klaus Schulten.
- Journal of Computational Chemistry, 28:2618-2640, 2007.
- Online full text of paper
- Online full text (JCC web site): http://dx.doi.org/10.1002/jcc.20829
- Journal of Computational Chemistry, 28:2618-2640, 2007.
- "A System for Interactive Molecular Dynamics Simulation"
- John E. Stone, Justin Gullingsrud, Klaus Schulten, and Paul Grayson.
- In 2001 ACM Symposium on Interactive 3D Graphics,
- John F. Hughes and Carlo H. Sequin, editors, pages 191-194, New York, 2001, ACM SIGGRAPH
- ACM Digital Library, citations, online full text, etc
- Online full text: http://doi.acm.org/10.1145/364338.364398
- In 2001 ACM Symposium on Interactive 3D Graphics,
- "An Efficient Library for Parallel Ray Tracing and Animation"
- John E. Stone
- Master's Thesis, University of Missouri-Rolla, Department of Computer Science, April 1998
- Master's Thesis, University of Missouri-Rolla, Department of Computer Science, April 1998
- "Rendering of Numerical Flow Simulations Using MPI"
- John Stone and Mark Underwood.
- Second MPI Developers Conference, pages 138-141, 1996.
- Online full text: http://dx.doi.org/10.1109/MPIDC.1996.534105
- Second MPI Developers Conference, pages 138-141, 1996.
- "Numerical Flow Simulation and Rendering Using MPI"
- John Stone, Mark Underwood
- In Proceedings of the 1996 Intel Supercomputer Users Group Conference
- Oak Ridge National Laboratory, Knoxville TN
- In Proceedings of the 1996 Intel Supercomputer Users Group Conference
- "An Efficient Library for Parallel Ray Tracing and Animation"
- John Stone
- In Proceedings of the 1995 Intel Supercomputer Users Group Conference
- Sandia National Laboratory, Albequerque NM
- In Proceedings of the 1995 Intel Supercomputer Users Group Conference
Short Articles, Blogs, and Whitepapers:
- "HPC Visualization on NVIDIA Tesla GPUs"
- John Stone
- NVIDIA Parallel ForAll Developer Blog, March 12, 2015
- NVIDIA Parallel ForAll Developer Blog, March 12, 2015
- "Random Numbers, Efficiency, and Other Things"
- John Stone
- Ray Tracing News, Volume 20, Number 1, May 19, 2007
- Ray Tracing News, Volume 20, Number 1, May 19, 2007
- "Real-Time GPU Spheres"
- John Stone
- Ray Tracing News, Volume 18, Number 1, December 12, 2005
- Ray Tracing News, Volume 18, Number 1, December 12, 2005
- "Workstation Clusters for Parallel Computing"
- John Stone, Fikret Ercal
- IEEE Potentials pages 31-33, April/May 2001
- IEEE Home page
- IEEE Potentials pages 31-33, April/May 2001
- "The Ups and Downs of Multithreaded Ray Tracing and Optimization"
- John Stone
- Ray Tracing News, Volume 12, Number 2, December 21, 1999
- Ray Tracing News, Volume 12, Number 2, December 21, 1999
- "Accelerating Software-Based MPEG Encoding Using the VIS Instruction Set"
- John Stone, Ahmad Zandi
- Interactive Insights: A Multimedia Compendium, pages 30-34,
- 1997 National Association of Broadcasters Convention
- Las Vegas, Nevada
- Interactive Insights: A Multimedia Compendium, pages 30-34,
Presentations:
- Opportunities to Use Advanced Rendering Methods to Improve Scientific Insight, Scientific Visualization Interest Group, HHMI Janelia Research Campus, Virtual (5/12/2022)
- The Future of GPU Ray Tracing, Panel Session, GPU Technology Conference, Virtual (3/22/2022)
- Full Time Interactive Ray Tracing for Scientific Visualization, From Atoms to the COVID-19 Virion, GPU Technology Conference, Virtual (3/22/2022)
- Experiences with Adopting ANARI in Existing Visualization Applications, Khronos Group ANARI webinar (3/2/2022)
- #COVIDisAirborne: AI-Enabled Multiscale Computational Microscopy of Delta SARS-CoV-2 in a Respiratory Aerosol, Supercomputing'21 ACM Gordon Bell COVID-19 Prize Finalist (11/17/2021)
- Intelligent Resolution: Integrating Cryo-EM with AI-Driven Multi-Resolution Simulations to Observe the SARS-CoV-2 Replication-Transcription Machinery in Action, Supercomputing'21 ACM Gordon Bell COVID-19 Prize Finalist (11/17/2021)
- ANARI Analytic Rendering Interface and VMD, NCSA Satellite Lightning Talks, IEEE Visualization '21 (10/26/2021)
- Using Next-Generation GPUs for Scientific Computing, Analysis, and Visualization, CECAM Extended Software Development Workshop: High performance computing for simulation of complex phenomena, Virtual (10/14/2021)
- Scalable Molecular Visualization and Analysis Tools in VMD, Argonne Training Program for Exascale Computing, Virtual (8/9/2021)
- Molecular Modeling, Visualization, and Analysis in VMD, MMBioS Computational Biophysics Workshop, Virtual (6/30/2021)
- Using Performance Analysis Tools to Speed Up Your Codes and Workflows, UIUC New Frontiers Initiative, National Geospatial Intelligence Agency, Webinar (6/1/2021)
- Full-Time Interactive Ray Tracing for Scientific Visualization: The VMD Experience, GPU Technology Conference, Virtual (4/15/2021)
- Cinematic Scientific Visualization: Challenges and Technological Opportunities, GPU Technology Conference, Virtual (4/15/2021)
- AI-Driven Multiscale Simulations Illuminate Mechanisms of SARS-CoV-2 Spike Dynamics, Supercomputing'20 ACM Gordon Bell COVID-19 Prize Winner (11/19/2020)
- NAMD and VMD Performance on ARM GPU Platforms, AHUG Supercomputing'20 (11/4/2020)
- Molecular Modeling, Visualization, and Analysis in VMD, MMBioS Computational Biophysics Workshop, Virtual (10/14/2020)
- Khronos ANARI: What's Happening Inside the ANARI Working Group and How YOU Can Get Involved!", Khronos Group Webinar (8/2020)
- Scalable Molecular Visualization and Analysis Tools in VMD, Argonne Training Program for Exascale Computing, Virtual (8/2020)
- Optimizing CUDA Kernels in HPC Simulation and Visualization Codes using NVIDIA Nsight Compute, GPU Technology Conference, Virtual (3/2020)
- Hybrid Molecular Mechanics/Artificial Intelligence Simulation Methods to Study Molecular Systems, GPU Technology Conference, Virtual (3/2020)
- Reduced Precision for Molecular Dynamics Simulation, Visualization, and Analysis, Reduced Precision Working Group, Webinar (2/2020)
- VMD: Preparation, Analysis, and Visualization of Molecular Dynamics Simulations, SBGrid Webinar (1/28/2020)
- SOLAR Consortium BoF on Accelerated Ray Tracing for Scientific Simulations, Supercomputing 2019, Denver, CO (11/20/2019)
- An Accessible Visual Narrative for the Primary Energy Source of Life from the Fulldome Show Birth of Planet Earth, Visualization and Data Analytics Showcase, Supercomputing 2019, Denver, CO (11/20/2019)
- NVIDIA CEO Special Address, Sheraton Denver Towntown, Denver, CO (11/18/2019)
- Initial NAMD and VMD Performance Results on ORNL Wombat, ORNL-NVIDIA ARM Project Meeting, Webinar (11/11/2019)
- Interoperability of Molecular Dynamics Visualization and Analysis Software, MolSSI Workshop on Molecular Dynamics Software Interoperability, New York, NY (11/4/2019)
- NCSA Blue Waters Petascale Computing Hackathon, NCSA, University of Illinois at Urbana-Champaign (09/09/2019 to 09/13/2019)
- NCSA Petascale Computing Institute, NCSA, University of Illinois at Urbana-Champaign (08/19/2019 to 8/23/2019)
- NAMD Developer's Workshop, Beckman Instititute, University of Illinois at Urbana-Champaign (08/19/2019 to 8/20/2019)
- Scalable Molecular Visualization with VMD, Argonne Exascale Training Program, Q Center, St. Charles, IL (08/05/2019)
- Birth of Planet Earth, Siggraph Computer Animation Festival, Los Angeles, CA (07/28/2019 to 8/3/2019)
- Interactive GPU Ray Tracing for Molecular Visualization, NVIDIA Exhibition, Siggraph, Los Angeles, CA (07/29/2019)
- Omnidirectional Stereoscopic Projections for VR, BoF on Immersive Visualisation for Science, Research, and Art, Siggraph, Los Angeles, CA (07/29/2019)
- Immersive Visualization and Interactive HPC Analytics for Analysis of Molecular and Cellular Simulations, Immersive Visualization Workshop, Siggraph, Los Angeles, CA (07/28/2019)
- 2019 PRACE HPC Summer School on Biomolecular Simulation, KTH, Stockholm, Sweden (06/10/2019 to 6/14/2019)
- Bringing State-of-the-Art GPU Accelerated Molecular Modeling Tools to the Research Community, GPU Technology Conference, San Jose, CA (03/20/2019)
- Interactive High Fidelity Biomolecular and Cellular Visualization with RTX Ray Tracing APIs, GPU Technology Conference, San Jose, CA (03/20/2019)
- Molecular Visualization and Simulation in VR, Indiana University, IN (3/6/2019)
- Adapting Scientific Software and Designing Algorithms for Next Generation GPU Computing Platforms, SIAM Conference on Computational Science and Engineering, Spokane, WA (02/27/2019)
- Challenges for Analysis and Visualization of Atomic-Detail Simulations of Minimal Cells, SIAM Conference on Computational Science and Engineering, Spokane, WA (02/26/2019)
- Optimizing NAMD and VMD for the IBM Power9 Platform, IBM Power User's Group, Supercomputing 2018, Dallas, TX (11/15/2018)
- NVIDIA HPC and University Computing Panel, Supercomputing 2018, Dallas, TX (11/14/2018)
- Using AWS EC2 GPU Instances for Computational Microscopy and Biomolecular Simulation, Amazon AWS Theater, Supercomputing 2018, Dallas, TX (11/14/2018)
- Interactivity in HPC BoF, Supercomputing 2018, Dallas, TX (11/13/2018)
- Putting High-Octane GPU-Accelerated Molecular Modeling Tools in the Hands of Scientists, NVIDIA Theater, Supercomputing 2018, Dallas, TX (11/13/2018)
- Blue Waters GPU Hackathon, National Center for Supercomputing Applications, University of Illinois, Urbana, IL (9/10/2018)
- GPU-Accelerated OptiX Ray Tracing for Scientific Visualization, NVIDIA Theater, Siggraph 2018, Vancouver BC, Canada (8/15/2018)
- VMD: Immersive Molecular Visualization with High-Fidelity Ray Tracing, Immersive Visualization for Science and Research, Siggraph 2018, Vancouver BC, Canada (8/13/2018)
- Argonne Training Program on Exascale Computing, Q Center, St. Charles, IL (8/9/2018)
- Preparing and Analyzing Large Molecular Simulations with VMD, Modeling Supra-molecular Structures with LAMMPS, Temple University, Philadelphia, PA (7/9 to 7/13, 2018)
- Interactive HPC Requirements, Challenges, and Solutions for Cutting Edge Molecular Simulation Science Campaigns, First Workshop on Interactive High-Performance Computing, ISC 2018, Frankfurt, Germany (6/28/2018)
- Visualizing the Atomic Detail Dynamics of Biomolecular Complexes in Our Compute-Rich but I/O-Constrained Future, Visualization in HPC Focus Session, ISC 2018, Frankfurt, Germany (6/25/2018)
- Keynote: Visualization Challenges and Opportunities Posed by Petascale Molecular Dynamics Simulations, Workshop on Molecular Graphics and Visual Analysis of Molecular Data, EuroVis 2018, Brno, Czech Republic (6/4/2018)
- Advances in Biomolecular Simulation with NAMD and VMD, Pawsey Supercomputing Centre, Perth, Australia (4/19/2018)
- Pawsey GPU Hackathon, Pawsey Supercomputing Centre, Perth, Australia (4/16 to 4/20, 2018)
- VMD: Biomolecular Visualization from Atoms to Cells Using Ray Tracing, Rasterization, and VR, GPU Technology Conference, San Jose, CA (3/29/2018)
- Using Accelerator Directives to Adapt Science Applications for State-of-the-Art HPC Architectures, GPU Technology Conference, San Jose, CA (3/27/2018)
- Accelerating Molecular Modeling on Desktop and Pre-Exascale Supercomputers, GPU Technology Conference, San Jose, CA (3/26/2018)
- Optimizing HPC Simulation and Visualization Code Using NVIDIA Nsight Systems, GPU Technology Conference, San Jose, CA (3/29/2018)
- A Brief History of How We Got Today's State-of-the-Art GPUs and What They Can Do, Illini E-Sports Student Organization, Digital Computer Laboratory, U. Illinois (1/27/2018)
- Interactive Supercomputing for State-of-the-Art Biomolecular Simulation, Interactivity in Supercomputing Birds of Feather, Supercomputing 2017, Denver, CO (11/14/2017)
- Using Accelerator Directives to Adapt Science Applications for State-of-the-Art HPC Architectures, Keynote Presentation, WAACPD'17: Fourth Workshop on Accelerator Programming Using Directives, Supercomputing 2017, Denver, CO (11/13/2017)
- Scalable Molecular Visualization and Analysis Tools in VMD, Argonne Training Program on Exascale Computing, Q Center, St. Charles, IL (8/10/2017)
- Communicating Science Through Visualization in the Age of Alternative Facts, Siggraph 2017 Courses, Los Angeles, CA (7/31/2017)
- Scaling to Petascale Institute, GPU Architecture Concepts and CUDA Programming, National Center for Supercomputing Applications, University of Illinois (6/28/2017)
- VMD Adaptation for HiHAT Proof of Concept Implementations, Modelado HiHAT meeting (remote webinar presentation) (06/20/2017)
- Brookhaven GPU Hackathon, Brookhaven National Laboratory, NY (6/5/2017 to 6/9/2017)
- VMD: GPU-Accelerated Analysis of Biomolecular and Cellular Simulations, Cray Analytics Symposium (remote webinar presentation), Radisson Blu MOA, Bloomington, MN (05/26/2017)
- GPUs Unleashed: Analysis of Petascale Molecular Dynamics Simulations with VMD, GPU Technology Conference, San Jose, CA (05/10/2017)
- Cutting Edge OptiX Ray Tracing Techniques for Visualization of Biomolecular and Cellular Simulations in VMD, GPU Technology Conference, San Jose, CA (05/09/2017)
- Turbocharging VMD Molecular Visualizations with State-of-the-Art Rendering and VR Technologies, GPU Technology Conference, San Jose, CA (05/09/2017)
- Visualizing Biomolecules in VMD, Hands-on Workshop on Computational Biophysics, NIH Center for Macromolecular Modeling and Bioinformatics, Beckman Institute, University of Illinois, Urbana, IL (4/18/2017).
- VMD: Preparation and Analysis of Molecular and Cellular Simulations, HPC for Life Sciences PRACE Spring School 2017, Stockholm Sweden (4/10/2017).
- Harnessing GPUs to Probe Biomolecular Machines at Atomic Detail, NVIDIA GPU Technology Theater, Supercomputing 2016, Supercomputing 2016, Salt Lake City, UT (11/16/2016).
- Visualization and Analysis of Biomolecular Complexes on Upcoming KNL-based HPC Systems: TACC Stampede-2 and ANL Aurora, Intel HPC Developers Conference, Supercomputing 2016, Salt Lake City, UT (11/13/2016).
- Immersive Molecular Visualization with Omnidirectional Stereoscopic Ray Tracing and Remote Rendering, High Performance Data Analysis and Visualization Workshop, Chicago (5/23/2016)
- High Performance Molecular Visualization: In-Situ and Parallel Rendering with EGL, High Performance Data Analysis and Visualization Workshop, Chicago (5/23/2016)
- Evaluation of Emerging Energy-Efficient Heterogeneous Computing Platforms for Biomolecular and Cellular Simulation Workloads, 25th International Heterogeneity in Computing Workshop, Chicago (5/23/2016)
- VMD+OptiX: Streaming Interactive Ray Tracing from Remote GPU Clusters to Your VR Headset, GPU Technology Conference, San Jose, CA (4/06/2016)
- VMD: Interactive Molecular Ray Tracing with NVIDIA OptiX, GPU Technology Conference, San Jose, CA (4/06/2016)
- VMD: Petascale Molecular Visualization and Analysis with Remote Video Streaming, GPU Technology Conference, San Jose, CA (4/05/2016)
- VMD: enabling preparation, visualization, and analysis of petascale and pre-exascale molecular dynamics simulations, Armadillo B453 R1001, Lawrence Livermore National Laboratory, Livermore, CA (1/5/2016)
- VMD+OptiX: Bringing Interactive Molecular Ray Tracing from Remote GPU Clusters to Your VR Headset, NVIDIA Theater, Supercomputing 2015, Austin, TX (11/18/2015)
- Chemical Visualization of Human Pathogens : The Retroviral Capsids, Visualization and Data Analytics Showcase, Supercomputing 2015, Austin, TX (11/18/2015)
- Frontiers of Molecular Visualization: Interactive Ray Tracing, Panoramic Displays, VR HMDs, and Remote Visualization, VisTech Workshop, Supercomputing 2015, Austin, TX (11/15/2015)
- Visualizing Biomolecular Complexes on x86 and KNL Platforms: Integrating VMD and OSPRay, Intel HPC Developer Conference, Austin TX (11/14/2015)
- VMD: Interactive Publication-Quality Molecular Ray Tracing with OptiX, NVIDIA Best of GTC Theater, Siggraph 2015, Los Angeles, CA (8/12/2015)
- VMD: Immersive Molecular Visualization and Interactive Ray Tracing for Domes, Panoramic Theaters, and Head Mounted Displays, BOF: Immersive Visualization for Science and Research, Siggraph 2015, Los Angeles, CA (8/10/2015)
- Workshop on Accelerated High-Performance Computing in Computational Sciences (SMR 2760), Abdus Salam International Centre for Theoretical Physics, Miramare - Trieste, Italy (05/27/2015)
- Panel: Data Relevancy, Locality, Diversity & More, NCSA Private Sector Program Annual Meeting, (05/06/2015)
- GPGPU 2015 Advanced Methods for Computing with CUDA, University of Cape Town, South Africa (4/20/2015)
- Proteins and Mesoscale Data: Visualization of Molecular Dynamics, Visualizing Biological Data (VIZBI) 2015 Conference, Broad Institute of MIT and Harvard, Cambridge, MA (03/26/2015)
- Publication-Quality Ray Tracing of Molecular Graphics with OptiX, GPU Technology Conference (03/19/2015)
- Guest Presentation: Integrating OptiX in VMD (in David McAllister's Innovations in OptiX presentation), GPU Technology Conference (03/18/2015)
- VMD: Visualization and Analysis of Biomolecular Complexes with GPU Computing, GPU Technology Conference (03/18/2015)
- Visualization of Energy Conversion Processes in a Light Harvesting Organelle at Atomic Detail, Visualization and Data Analytics Showcase, Supercomputing 2014, New Orleans, LA (11/19/2014)
- GPU-Accelerated Analysis of Large Biomolecular Complexes, NVIDIA Booth, Supercomputing 2014, New Orleans, LA (11/18/2014)
- GPU-accelerated Visualization and Analysis of Petascale Molecular Dynamics Simulations, Integrated Imaging Initiative Workshop: Tomography and Ptychography, Argonne National Laboratory (9/30/2014)
- GPU-accelerated Visualization and Analysis of Petascale Molecular Dynamics Simulations, Research and Technology Development Conference, Missouri University of Science and Technology (9/16/2014)
- Programming for Hybrid Architectures Today and in the Future, LSU Third Annual HPC User Symposium, Louisiana State University (6/5/2014)
- GPU-Accelerated Visualization and Analysis of Biomolecular Complexes, Oxford University, United Kingdom (5/12/2014)
- GPU-Accelerated Analysis and Visualization of Large Structures Solved by Molecular Dynamics Flexible Fitting, Faraday Discussion 169: Molecular Simulations and Visualization, University of Nottingham, United Kingdom (5/8/2014)
- GPGPU2 Advanced Methods for Computing with CUDA, University of Cape Town, South Africa (4/22/2014)
- Petascale Molecular Ray Tracing: Accelerating VMD/Tachyon with OptiX, GPU Technology Conference (03/27/2014)
- Fighting HIV with CUDA, CCoE Achievement Award, GPU Technology conference (03/25/2014)
- Visualization and analysis of petascale molecular simulations with VMD, GPU Technology Conference (03/25/2014)
- Using GPUs to Supercharge Visualization and Analysis of Molecular Dynamics Simulations with VMD, NVIDIA GTC Express (02/25/2014)
- Experiences Developing and Maintaining Scientific Applications on GPU-Accelerated Platforms, Indiana University, IUPUI Campus, Indianapolis, IN (01/24/2014)
- Fighting HIV with GPU-Accelerated Petascale Computing, Supercomputing 2013, Denver, CO (11/19/2013)
- GPU-Accelerated Molecular Visualization on Petascale Supercomputing Platforms, UltraVis'13: Eighth Ultrascale Visualization Workshop, Denver, CO (11/17/2013)
- Visualization and Analysis of Petascale Molecular Dynamics Simulations, San Diego Supercomputer Center, University of California San Diego (11/5/2013)
- Experiences Developing and Maintaining Scientific Applications on GPU-Accelerated Platforms, Indiana University, Bloomington, IN (10/2/2013)
- Interactive Molecular Visualization and Analysis with GPU Computing, Fall National Meeting of the American Chemical Society, Indianapolis, IN (9/11/2013)
- Early Experiences Scaling VMD Molecular Visualization and Analysis Jobs on Blue Waters, XSEDE Extreme Scaling Workshop, Boulder, CO (8/15/2013)
- GPU-Accelerated Molecular Visualization and Analysis with VMD, Midwest Theoretical Chemistry Conference, Urbana, IL, (5/31/2013)
- GPGPU Computing with CUDA Workshop, University of Cape Town, South Africa (4/29/2013)
- VMD: GPU-Accelerated Visualization and Analysis of Petascale Molecular Dynamics Simulations, GPU Technology Conference, San Jose, CA (3/20/2013)
- Keynote: Broadening the Use of Scalable Kernels in NAMD/VMD, VSCSE Many-core Processors, NCSA, University of Illinois (8/16/2012)
- GPU-Accelerated Analysis of Petascale Molecular Dynamics Simulations with VMD, Scalable Software for Scientific Computing Workshop, University of Notre Dame (6/11/2012)
- Visualization of petascale molecular dynamics simulations, Imaging at Illinois, The Next Generation: Computational Imaging and Visualization, Beckman Institute, University of Illinois (6/1/2012)
- In-Situ Visualization and Analysis of Petascale Molecular Dynamics Simulations with VMD, Accelerated HPC Symposium, San Jose, CA (5/17/2012)
- High Performance Molecular Visualization and Analysis on GPUs, GPU Technology Conference, San Jose, CA (5/16/2012)
- Immersive Out-of-Core Visualization of Large-Size and Long-Timescale Molecular Dynamics Trajectories, ISVC 2011, Las Vegas, NV (9/26/2011)
- Immersive Molecular Visualization and Interactive Modeling with Commodity Hardware, ISVC 2010, Las Vegas, NV (12/1/2010)
- Faster, Cheaper, Better: Biomolecular Simulation with NAMD, VMD, and CUDA, NVIDIA Booth, Supercomputing 2010, New Orleans, LA (11/16/2010)
- High Performance Computing with CUDA Case Study: Heterogeneous GPU Computing for Molecular Modeling, CUDA Tutorial, Supercomputing 2010, New Orleans, LA (11/14/2010)
- High Performance Molecular Simulation, Visualization, and Analysis on GPUs, GPU Technology Conference (09/22/2010)
- High Performance Molecular Simulation, Visualization, and Analysis on GPUs, Oak Ridge National Laboratory (09/16/2010)
- Faster, Cheaper, and Better Science: Molecular modeling on GPUs, Fall National Meeting of the Americal Chemical Society, Boston, MA (8/22/2010)
- OpenCL: Molecular modeling on heterogeneous computing systems, Fall National Meeting of the Americal Chemical Society, Boston, MA (8/22/2010)
- Lecture 4: Cut-off and Binning for Regular Data Sets, VSCSE Many-core Processors, University of Illinois (8/03/2010)
- The OpenCL Programming Model, Part 1 Illinois UPCRC Summer School (7/23/2010)
- The OpenCL Programming Model, Part 2 Illinois UPCRC Summer School (7/23/2010)
- Molecular Visualization and Analysis on GPUs, Symposium on Application of GPUs in Chemistry and Materials Science, University of Pittsburgh (06/29/2010)
- ECE498AL Application Performance Case Studies: Molecular Visualization and Analysis, University of Illinois at Urbana-Champaign (04/08/2010)
- An Introduction to OpenCL, GPUComputing.net Webinar (12/10/2009)
- OpenCL for Molecular Modeling Applications: Early Experiences, OpenCL BOF, Supercomputing 2009, Portland, OR (11/18/2009)
- Accelerating Molecular Modeling Applications with GPU Computing, Exhibition, Supercomputing 2009, Portland, OR (11/18/2009)
- High Performance Computing with CUDA Case Study: Molecular Modeling Applications,CUDA Tutorial, Supercomputing 2009, Portland, OR (11/15/2009)
- An Introduction to OpenCL, IACAT/CCOE GPU Brown Bag Forum, University of Illinois (10/21/2009)
- High Performance Molecular Visualization and Analysis with GPU Computing, Beckman Institute Forum for Imaging and Visualization, University of Illinois (10/20/2009)
- GPU Accelerated Visualization and Analysis in VMD and Recent NAMD Developments, GPU Technology Conference, San Jose, CA (10/1/2009)
- Case Study - Accelerating Molecular Dynamics Experimentation, VSCSE: Many-Core Processors for Science and Engineering Applications, NCSA (8/13/2009)
- Multidisciplinary Panel, VSCSE: Many-Core Processors for Science and Engineering Applications, NCSA (8/10/2009)
- Accelerating Molecular Dynamics on a GPU, Careers in High-Performance Systems Mentoring Workshop, National Center for Supercomputing Applications (7/25/2009)
- GPU Accelerated Visualization and Analysis in VMD, Center for Molecular Modeling, University of Pennsylvania (6/9/2009)
- Keynote: Accelerating Molecular Modeling Applications with GPU Computing, Second Sharcnet Symposium on GPU and Cell Computing, University of Waterloo (5/20/2009)
- ECE498AL Application Performance Case Studies: Molecular Visualization and Analysis, University of Illinois at Urbana-Champaign (04/09/2009)
- Experiences with Multi-GPU Acceleration in VMD, Path to Petascale: Adapting GEO/CHEM/ASTRO Applications for Accelerators and Accelerator Clusters, NCSA (4/2/2009)
- High Performance Computation and Interactive Display of Molecular Orbitals on GPUs and Multi-core CPUs, Second Workshop on General-Purpose Processing on Graphics Processing Units, Washington D.C. (3/8/2009)
- High Performance Computation and Interactive Display of Molecular Orbitals on GPUs and Multi-core CPUs, IACAT Accelerator Workshop, NCSA (1/23/2009)
- GPU Computing Case Study: Molecular Modeling Applications, ECE 598SP: Massively Parallel Processors, University of Illinois (11/11/2008)
- GPU Computing, Cape Linux Users Group, South Africa (10/28/2008)
- Visualizing Biomolecular Complexes with VMD, Centre for High Performance Computing, CSIR Rosebank Campus, Cape Town, South Africa (10/27/2008)
- An Introduction to Molecular Visualization with VMD, Centre for High Performance Computing, CSIR Rosebank Campus, Cape Town, South Africa (10/27/2008)
- Accelerating Molecular Modeling Applications with Graphics Processors, Computer Science Department, University of Cape Town, South Africa (10/23/2008)
- Accelerating Computational Biology by 100x Using CUDA, NVISION 08, San Jose Convention Center, San Jose, CA (8/26/2008)
- Case Study - Accelerating Molecular Dynamics Experimentation, VSCSE: Accelerators for Science and Engineering Applications: GPUs and Multicore, NCSA (8/21/2008)
- Accelerating Scientific Applications with GPUs, Workshop on Programming Massively Parallel Processors, NCSA (7/10/2008)
- Accelerating Molecular Modeling Applications with Graphics Processors, SIAM conference on Parallel Processing for Scientific Computing, Atlanta, GA (3/12/2008)
- Case Study: Molecular Visualization and Analysis, Supercomputing 2007 Tutorial: High Performance Computing with CUDA, Reno, NV (11/11/2007)
- GPU Acceleration of Scientific Applications Using CUDA, AstroGPU 2007, Institute for Advanced Study, Princeton NJ (11/09/2007)
- ECE498AL CUDA Performance Case Studies: Ion Placement Tool, VMD, University of Illinois at Urbana-Champaign (10/15/2007)
- ECE498AL CUDA Performance Case Studies: Ion Placement Tool, VMD, University of Illinois at Urbana-Champaign, (3/14/2007)
- Visualization of Nano-Scale Structures, University of Texas Health Science Center at Houston (4/20/2006)
- Universidad Nacional Autónoma de Mexico, DGSCA (8/11/2006) (Video Conference Presentation)
- VMD: Algorithms and Methods for Large Scale Biomolecular Visualization, San Diego Supercomputer Center (9/12/2005)
- Dancing Proteins: 3-D Visualization of Protein Structure and Dynamics on Next-Generation Graphics Hardware, ITG Forum at University of Illinois at Urbana-Champaign (2/15/2005)
- CS Colloqium at University of Missouri-Rolla (2/22/2005)
- Purdue University Envision Center (2/25/2005)
- VMD: Biomolecular Visualization and Analysis, The Scripps Research Institute, La Jolla CA (7/31/2003)
- Designing a Cluster for a Small Research Group, Summer School on Theoretical Biophysics, Beckman Institute UIUC, 6/11/2003
- Biomolecular Visualization in the CAVE with VMD, Center for Parallel Computing, Swedish Royal Institute of Technology, Stockholm Sweden (10/3/2001)
- Cluster-based visualization with VMD on a tiled display wall, NCSA Terascale Clusters Dedication (9/5/2001)
- Biomolecular Visualization, Interactive Molecular Dynamics, Siggraph 2001, Sun Microsystems Booth (8/14/2001)
- Low-Cost Linux Clusters for Biomolecular Simulations Using NAMD, Beckman Institute UIUC, June 26, 2001
- Software Development Tutorial for Biomedical Research Scientists, Beckman Institute UIUC, April 3, 2001
- Linux Clusters, Beckman Institute UIUC, November 11, 1999
Service:
- Editorial Board Member, Journal of Molecular Graphics and Modelling (2010-2013)
- Guest Editorial Board Member, IEEE TPDS Special Issue on High Performance Computing with Accelerators (2009)
- Interactive High Performance Computing (InteractiveHPC) (2018, ISC-2019, SC-2019)
- International Conference on Algorithms and Architectures for Parallel Processing (ICA3PP) (2017)
- Euro-Par (2016)
- International workshop on OpenCL (IWOCL) (2013, 2014, 2015)
- Sixth Workshop on General Purpose Processing on Graphics Processing Units (2013)
- Workshop on Programmability Issues for Heterogeneous Multicores (MULTIPROG 2011, 2012, 2013)
- Symposium on Application Accelerators in High Performance Computing (SAAHPC 2009, 2010, 2011, 2012)
- IEEE International Parallel & Distributed Processing Symposium (IPDPS 2011)
- International Symposium on Visual Computing (ST3 2011)
- IEEE International Workshop on High Performance Computational Biology (HiCOMB 2009, 2010)
- Workshop on Language, Compiler, and Architecture support for GPGPU (2010)
- NCSA/IACAT Workshop on Programming Massively Parallel Processors (PMPP 2008)
- Programming Massively Parallel Processors: A Hands-on Approach, Third Edition (chapter reviewer, published 2016)
- GPU Computing Gems Emerald Edition (chapter reviewer, published 2011)
- GPU Gems 3 (chapter reviewer, published 2007)
- Geometric Tools for Computer Graphics (reviewer, published 2002)
- Real-Time Rendering (reviewer, first edition, published 1999)
- Siggraph (2008, 2020, 2022)
- EuroVis (2014, 2015, 2019, 2020, 2022)
- IEEE VR (2017, 2021)
- Journal of Computational Physics (2018, 2019)
- Computers and Geosciences (2018, 2019)
- IEEE Transactions on Computers (2017, 2018)
- Concurrency and Computation: Practice and Experience (2017)
- Euro-Par (2012, 2016)
- Algorithms (2016)
- IEEE Pacific Visualization (2015)
- Computer Graphics Forum (2008, 2009, 2015)
- PLoS Computational Biology (2015)
- IEEE Transactions on Parallel and Distributed Systems (2014)
- International Workshop on OpenCL (IWOCL) (2013, 2014)
- Journal of Molecular Graphics and Modelling (2013)
- Workshop on General Purpose Processing on GPUs (2013)
- Computing in Science and Engineering (2009, 2011, 2012)
- PLoS ONE (2012)
- Supercomputing (2009, 2011)
- Parallel Computing, (2010, 2011)
- Computing Frontiers (2011)
- High Performance Graphics (2010)
- Computer Physics Communications (2010)
- Journal of Computational Chemistry (2008, 2009, 2010)
- Computers and Graphics (2010)
- Visualization in Medicine and Life Sciences (2009)
- PPAM 2009 Eighth International Conference on Parallel Processing and Applied Mathematics (2009)
- BMC Structural Biology (2008, 2009)
- Journal of Parallel Computing (2008)
- IEEE VIS (2007)
- 2006 IEEE Symposium on Interactive Ray Tracing
- ACM Computing Surveys (2006)
- Virtual Reality, Volume 9 number 1 (2005)
- International Journal of Parallel and Distributed Systems and Networks (2002)
- IEEE International Parallel & Distributed Processing Symposium (2000)
- DOE Exascale Computing Project - Software Technology Proposals (2016)
- Computer Science Advisory Board, Missouri University of Science and Technology (2007-2013, 2019)
- Biomedical Analysis and Simulation Supercomputer Advisory Board, NIH Resource for Computer-Integrated Systems for Microscopy and Manipulation (CISMM), University of North Carolina at Chapel Hill (2009, 2010)
- GPU-Accelerated Clustering Analysis of Molecular Dynamics Trajectories Using QCP and PAM (2015),
Devin King and Julie Xia - Utilizing Wide Vector Instructions with NAMD (2014),
Craig Disselkoen and Jordan Jorgensen - GPU-accelerated Brownian Dynamics (2013),
Justin Dufresne and Terrance Howard - GPU-accelerated Secondary Structure Analysis (2013),
Alex Knaust and John McCann - GPU-accelerated Molecular Dynamics Simulation Analysis (2012),
Jason Hutcheson and Michael Musson - GPU-accelerated Molecular Dynamics Simulation Analysis (2011),
Sean Karlage and Michael Robson
- Arjun Radhakrishnan, M.Sc. Electrical Engineering, "Accelerating Pulsar Dedispersion for MeerKAT on Graphics Processing Units", University of Cape Town, South Africa, 2010
- Juan-Pierre Longmore, M.Sc. Computer Science, "Towards Realistic Interactive Sand: A GPU-based Framework", University of Cape Town, South Africa, 2009
- Judging panel for SCS Science Image Challenge competitions (2011, 2012, 2013, 2014)
Press Articles and Interviews:
- Nature Computational Science: Fighting COVID-19 with HPC, Dec 13, 2021
- New York Times: The Coronavirus in a Tiny Drop, Dec 1, 2021
- Seimaxim: Researchers Fight COVID, Advance Science with NVIDIA A100 GPU, Nov 23, 2021
- UCSD: COVID Gets Airborne, Nov 22, 2021 , Nov 23, 2021
- UCL: UCL and UK scientists part of team nominated for supercomputing prize, Nov 18, 2021
- EurekAlert: We know #COVIDisAirborne - Now we have the first ever model of an aerosolized viral particle, Nov 17, 2021
- Oak Ridge National Laboratory: Waltzing the virus: Study on COVID-19 reproduction earns Gordon Bell Special Prize nomination, Nov 17, 2021
- OLCF: Waltzing the virus: Study on COVID-19 reproduction earns Argonne researchers Gordon Bell Special Prize nomination, Nov 16, 2021
- Argonne National Laboratory: Waltzing the virus: Study on COVID-19 reproduction earns Argonne researchers Gordon Bell Special Prize nomination, Nov 16, 2021
- Charmworks: Two projects using Charm++ named finalists for Gordon Bell Special Prize, Nov 15, 2021
- HPCWire: Gordon Bell Finalists Fight COVID, Advance Science with NVIDIA Technologies, Nov 15, 2021
- NVIDIA: Gordon Bell Finalists Fight COVID, Advance Science with NVIDIA Technologies, Nov 15, 2021
- Krell CSGF: Team Including Fellow and Alumnus Wins HPC Prize for COVID-19 Research, Nov 20, 2020
- PHYS.ORG: Collaborative AI effort unraveling SARS-CoV-2 mysteries wins Gordon Bell Special Prize, Nov 20, 2020
- FederalLabs.org: DOE labs help their collaborative research teams nab two SC20 prizes, Nov 20, 2020
- Daily Herald: Argonne's coronavirus research earns Gordon Bell award, Nov 20, 2020
- sciencenewsnet.in: Gordon Bell Special Prize for COVID-19 Research Announced, Nov 20, 2020
- HPCWire: Gordon Bell Special Prize Goes to Massive SARS-CoV-2 Simulations, Nov 19, 2020
- InsideHPC: Winners of Student Cluster Competition, Gordon Bell Prize Named at SC20, Nov 19, 2020
- Argonne National Laboratory: Collaborative AI effort unraveling SARS-CoV-2 mysteries wins prestigious Gordon Bell Special Prize, Nov 19, 2020
- NVIDIA: COVID-19 Spurs Scientific Revolution in Drug Discovery with AI, Nov 19, 2020
- TACC: Gordon Bell Special Prize Winning Team Reveals AI Workflow for Molecular Systems In the Era of COVID-19, Nov 19, 2020
- UCSD: UC San Diego Leads Research that Earns Gordon Bell Special Prize, Nov 19, 2020
- EurekAlert: Collaborative AI effort unraveling SARS-CoV-2 mysteries wins Gordon Bell Special Prize, Nov 19, 2020
- Oak Ridge National Laboratory: Gordon Bell Special Prize Finalist Team Reveals AI Workflow for Molecular Systems in the Era of COVID-19, Nov 18, 2020
- HPCWire: AHUG at SC20: Arm HPC User Group Kicks off with 25 Presentations, Three Live Panels, Nov 5, 2020
- Next Platform: "Wombat" puts ARM's SVE instruction set to the test, Nov 18, 2020
- New York Times: The Coronavirus Unveiled, Oct 9, 2020
- Supercomputing 2019: From Atoms to Astronomy, HPC is Redefining Scientific Research, Feb 7, 2019
- IBM: 2019 Champions press release, Jan 16, 2019
- IBM: 2018 Champions press release, Jan 31, 2018
- IBM Systems: "Rock Stars in our Midst, by Eve Daniels, May 2017
- InsideHPC: "Rock Stars of HPC: John Stone", by Beth Harlen, InsideHPC, April 19, 2017
- IBM: 2017 Champions press release, Feb 3, 2017
- InsideHPC: "Visualizing Biomolecular Complexes on x86 and KNL Platforms: Integrating VMD and OSPRay", by Rich Brueckner, InsideHPC, December 18, 2015
- Scientific Computing World: "Does HPC's future lie in remote visualization", by Tom Wilkie, Scientific Computing World, February/March, 2015
- InsideHPC: "HPC's Future Lies in Remote Visualization, by Tom Wilkie, InsideHPC, February 11, 2015
- NVIDIA: "CUDA for ARM Platforms is Now Available, by Mark Harris, NVIDIA, June 18, 2013
- National Science Foundation: "Cracking the Code of HIV; Providing An Up-Close View of the Enemy", by Diana Yates, UIUC, and Lisa-Joy Zgorski NSF, May 29, 2013
- Inside HPC: "GPUs Helps Researchers Uncover New Approach to Combating HIV Virus", by Rich Brueckner, May 29, 2013
- PC Magazine: "Nvidia GPUs Assist in Battling HIV Virus", by Damon Poeter, May 29, 2013
- Bio-IT World: "Researchers Characterize Chemical Structure of HIV Capsid, by Allison Proffitt, May 29, 2013
- NVIDIA: "Breakthrough in HIV Research Enabled by NVIDIA GPU Accelerators", by George Millington, May 29, 2013
- Tech News Daily: "Supercomputer Tackles Mystery of HIV's Structure", by Jillian Scharr, May 29, 2013
- HPCwire: NVIDIA Releases CUDA 4.0 Toolkit, February 28, 2011
- University of Illinois: "Illinois Researcher Named 2010 CUDA Fellow by NVIDIA", by Laurie Talkington, December 13, 2010
- Scientific Computing: "Speeding Up Science", by Mike May, November 20, 2010
- NVIDIA: "NVIDIA Names Three New 2010 CUDA Fellows", November 10, 2010
- Chemical and Engineering News: "The GPU Revolution, by Lauren Wolf, November 1, 2010
- GenomeWeb: "Eyeing a Growing Market, Nvidia Launches Portal to Aggregate GPU-Enabled Life Science Applications", by Vivien Marx, January 15, 2010
- CNN/Money: "Tesla Bio Workbench Enables Scientists to Achieve New Breakthroughs in Biosciences", January 14, 2010
- SciDAC Review: "Hardware: Core Strength", Number 15, Winter 2009
- TG Daily: "NVIDIA teams up with Microsoft for HPC", by Andrew Thomas, September 28, 2009
- Wired: "Personal Supercomputers Promise Teraflops on Your Desk", by Priya Ganapati, August 3, 2009
- NVIDIA: "NVIDIA Appoints First CUDA Center of Excellence", Andrew Humber, June 30, 2008
- Bio-IT World: "Life Scientists Get Their Game Faces On", by Mike May, April 1, 2008
- Genome Technology: "Not Just for Kids Anymore", by Matthew Dublin, September, 2007
- BeHardware: "NVIDIA CUDA: practical uses" (English), by Damien Triolet, August 17, 2007
- Hardware.fr: "NVIDIA CUDA: plus en pratique" (French), by Damien Triolet, August 9, 2007
- TG Daily: "NCSA: A look inside one of the world's most capable supercomputer facilities", by Rick C. Hodgin, July 2, 2007
- Beyond3D: "NVIDIA Tesla: GPU computing gets its own brand", by Tim Murray, June 20, 2007
- TG Daily: "NVIDIA unveils Tesla, moves into supercomputing", by Wolfgang Gruener, June 20, 2007
Consulting Activities:
- "Graphics Processing Unit (GPU) Acceleration for Cosite Interference Prediction Tools", SBIR N131-008, funded by NAVAIR, U.S. Navy, 2013-2016.
- "Antenna Placement Optimization on Large, Airborne, Naval Platforms", SBIR N101-022, funded by NAVAIR, U.S. Navy, 2010-2013.
- "Multilevel Parallelization of Software for Accurate Protein-Ligand Affinities", SBIR PAR-07-160, 1R44GM088867, funded by the National Institutes of Health, 2010-2014.
Previous Employment:
- I was previously a Senior Programmer/Analyst at Heuris/Pulitzer, and later Heuris Logic, developing commercial video encoding and transcoding software used in service bureaus, the standalone commercial MPEG Power Professional software, and plug-ins for the leading non-linear video editing systems. My work in this era was noteworthy mainly for my early use of parallel processing and SIMD instruction set extensions for acceleration of the routines for color space conversions, image filtering and de-noising, and motion estimation within the video encoder subsystem.
Education:
- Master of Science in Computer Science
- Master's Program Advisor: Dr. Fikret Ercal
- University of Missouri-Rolla
- University of Missouri-Rolla
- Bachelor of Science in Computer Science
- University of Missouri-Rolla
Honors and Other Accomplishments:
- Winner of the ACM Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research for the paper entitled: "AI-Driven Multiscale Simulations Illuminate Mechanisms of SARS-CoV-2 Spike Dynamics"
- Winner of the SC'19 Visualization and Data Analytics Showcase for the movie and accompanying paper entitled: "An Accessible Visual Narrative for the Primary Energy Source of Life from the Fulldome Show Birth of Planet Earth"
-
IBM Champion for Power, 2017, 2018, 2019 - Finalist, 2016 GPU Center of Excellence Achievement Award: "Interactive Molecular Ray Tracing on GPU-Accelerated PCs, Remote Clouds and Supercomputers, and VR HMDs"
-
Our 2016 article Molecular dynamics-based refinement and validation for sub-5Å cryo-electron microscopy maps has received an F1000Prime recommendation 
- Finalist, 2015 NVIDIA Global Impact Award "Enabling Computational Virology with GPU Computing"
- Winner of the SC'14 Visualization and Data Analytics Showcase for the movie and accompanying paper entitled: "Visualization of Energy Conversion Processes in a Light Harvesting Organelle at Atomic Detail"
- Most Cited Article Since 2009, Journal of Molecular Graphics and Modelling
- 2014 CUDA Center of Excellence Achievement Award: Fighting HIV with CUDA
- NVIDIA CUDA Fellow: NVIDIA 2010 CUDA Fellows UIUC Press Release
- 2010 HiPEAC Paper Award for the paper "An Asymmetric Distributed Shared Memory Model for Heterogeneous Parallel Systems" presented at ASPLOS 2010.
- Tachyon, the ray tracing package I originally wrote for my Master's Thesis and continue to maintain today, was selected as a finalist for the SPEC MPI benchmark and is now part of the SPEC MPI2007 benchmark suite.
- Member in good standing of ACM, Siggraph, and IEEE.
- I hold an FCC Amateur Extra Class radio license KC9VGG, and a GMRS radio license. ARRL life member, and AMSAT-NA member. AMSAT OSCAR Satellite Communications Achievement Award #594, Limited Edition 50th Anniversary, March 6, 2019. ARRL WAS awards for Digital (JT65) #1900, and Mixed #60792, communication modes. WPX Digital award #983. ARISS SSTV award, Febaruary 2019.
- Member of the Champaign-Urbana Astronomical Society (CUAS). I have previously served as Vice President (2008, 2009, 2010), and Fifth Director (2013).
- 17th Place in 21st Annual ACM International Collegiate Programming Contest,
- 1996 World Finals, Philadelphia PA
- 1st Place in 1996 ACM Mid-Central Regional Programming Contest
- First Place Prize Winner: Intelligent Systems Center Graduate Research Paper Presentation Contest, University of Missouri-Rolla, April 20, 1996
- Prize Winner: Intelligent Systems Center Graduate Research Paper Presentation Contest, University of Missouri-Rolla, March 30, 1995
- Member of the Upsilon Pi Epsilon international computer science honorary society while a graduate student at University of Missouri-Rolla. I served as the treasurer for the local chapter in 1995.
- Member of the Kappa Kappa Psi national honorary band fraternity while a graduate student at University of Missouri-Rolla.
- Founding member of the University of Missouri-Rolla Solar Car Team while an undergraduate student.
- Member of the six man ISRA junior rifle team that won the 1990 National Trophy Team match at the National Trophy Rifle Matches at Camp Perry, Ohio.