Highlights of our Work
2026 | 2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
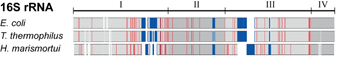
Life on Earth today is based on two fundamental classes of molecules, the molecules of the genetic
material, RNA and DNA, and the molecules of life's machines, proteins. At the dawn of life billions
of years ago, RNA apparently reigned pretty much by itself, DNA and proteins coming on board later. Eventually,
early living organisms began to specialize and then split into the three domains, Bacteria, Archaea, and Eucarya.
From there, life continued to evolve, branching out into the
many and diverse forms that surround us today. Life itself gave scientists the clock to study
this process of evolution. The clock is nothing less but the central machine of all living cells, the ribosome,
that reads genetic information and translates it into proteins. The first to read this clock and
establish the modern science of evolution was Carl Woese. The ribosome differs greatly from other
cellular machines in that it is made mainly of RNA, only to a smaller part of proteins, the latter
being added in the course of evolution. A recent report using the
MultiSeq tool of VMD
investigated the interaction between RNA and proteins in the ribosome. The researchers examined the
details of how RNA and proteins evolved together and showed how such details
can lead to better understanding of the ribosome as a machine. More details are available at the researchers'
web page.
The most powerful compute engine in your laptop and home computer is
what? The central processing unit, of course. Wrong, it is the
graphics processing unit (GPU)! Since GPUs are specialized for
graphics and games, popular uses of modern computers, their
development benefitted from strong market forces. Modern GPUs reach
Teraflop speed, passing entire computer clusters filling a room.
Unfortunately, GPUs lie dormant when computers are employed for
intensive biological computing like biomolecular simulations. GPUs
are now getting a wake-up call from biomedical researchers who then
enjoy many-fold speed-ups of their laptop computations
(see the Oct 2007 highlight),
vendors already now hawking GPU-powered "desktop
supercomputers." The next challenge is to bring the GPU from the
laptop to computer centers, speeding up the world's fastest
supercomputers. Building on previous development experience of
NAMD
and
VMD, a
recent report
describes NAMD running at impressive speed
on a GPU cluster. The report suggests techniques, useful also for
programmers of other applications to efficiently accelerate computer
clusters through GPUs. The report is particularly timely with a new,
large GPU-accelerated computer cluster coming on-line. The reported
GPU-based speed-ups will permit biomolecular simulations largely
unfeasible so far for studies of entire virus particles and cellular
organelles. More information here.

image size:
78.4KB
made with VMD
If you have a leaky roof, you need to plug the hole quickly to avoid further damage. Similarly, cells do not tolerate open holes in their surrounding
membranes for long. However, cell membranes do require channels for various molecules to get across, and proteins are no exception. The
protein-conducting channel SecY, a membrane-bound protein itself, is the pathway used by other proteins to cross the membrane. When it's not in use, it
needs to have a water-tight seal, which is provided by two elements in the channel, a bulky plug and a constrictive pore ring (see the April 2006 highlight). But do these two elements, plug and pore ring, function separately to provide two
independent barriers or together, providing a single barrier? Molecular dynamics simulations on the channel and two mutants in which a portion of the
plug was deleted have provided the answer, as reported in a recent publication: both
components are necessary to prevent leaks in the channel. More information on the amazing channel can be found on our Protein Translocation website.

image size:
661.9KB
made with VMD
Biological cells are surrounded by a highly versatile, yet very feeble cellular membrane and need to balance
differences between the cell's interior and exterior that otherwise would burst the membrane. For example, the osmotic
pressures inside and outside the cell need to be closely balanced. Thousands of proteins in the membrane act as gatekeepers,
opening pores that can also act as safety valves, helping to reduce the interior-exterior difference in pressure rapidly. One
such protein, the mechanosensitive channel of small conductance MscS (see the Mar 2008 highlight, "Observation and
Simulation Depict Cell's Safety Valve", the Feb
2007 highlight, "Observing and Modeling a Crucial Membrane Channel", the May 2006
highlight, "Electrical Safety Valve", and the Nov 2004
highlight, "Japanese Lantern Protein") opens in response to cellular membrane tension
generated due to a drastic imbalance in osmotic pressure as it arises when a bacterial cell suddenly finds itself in fresh
water, rather than a highly saline physiological medium. The MscS channel widens then to jettison molecules out of the
cell and reduce tension on the cellular membrane quickly. In a recent report, researchers have combined experimental data
from electron
paramagnetic measurements and computer modeling to reveal in atomic detail how MscS opens and closes its channel. Combining
measurement and modeling, the researchers established a highly resolving computational microscope, unmatched by existing
microscopes (more on our MscS website).

image size:
287.1KB
made with VMD
Living cells organize many functions: molecular import and export, signaling, transcription of genes into proteins, movement, building, repair, and more. These functions are realized through a complex architecture of the cell interior reflected in a system of labyrinthine membranes forming manifold cellular organelles from tubes, vesicles and many other shapes. Accordingly, cells need to sculpt their membrane in never ceasing processes and have at their disposal a wide range of mechanisms. A key sculpting mechanism is furnished by proteins, so-called BAR domains, that apparently form lattice-like scaffolds adhering to membrane surfaces. Such scaffolds have been observed through electron microscopy and they are now also being described through molecular dynamics simulations using NAMD. As recently reported, the simulations, carried out at four different levels of resolution (from an atomic to a continuum level), revealed that different arrangements of BAR domains lead to different curvatures. The simulations help to explain why BAR domains working in teams, i.e., in lattice formation, sculpt intra-cellular membranes into different shapes, depending on the exact arrangement. An arrangement of BAR domains that is particularly efficient in bending membranes was identified. More information can be found on our BAR domain web page.
image size:
360.9KB
made with VMD
Chromatophores are the photosynthetic machineries of bacteria. Each chromatophore contains, embedded in a membrane, all the photosynthetic proteins needed to absorb sunlight and turn it into chemical fuel. Chromatophores come in different shapes: while some chromatophores are spherical, others are flat or tubular. It has puzzled scientists how all these different geometries arise, and a hypothesis has developed that it is the photosynthetic proteins that render the shape of chromatophore membrane. In a study reported recently, computational biologists using NAMD took an atomistic look at how the chromatophore proteins bend the membrane. Simulations showed that the most numerous photosynthetic proteins dome the membrane, building arched membrane patches that can then be assembled into a spherical chromatophore. These simulations demonstrated that photosynthetic proteins construct their individual membrane environment, and when many of such proteins come together in the bacterial membrane, they can build functional cellular units with unique geometries. For more details, please see our chromatophore website.
Ouch!! You cut your finger with a knife. Blood immediately starts coming out from the wound, and in a panic you search through your drawer for a band-aid. However, well before you can find the band-aid, and even before the "ouch" came out, just about the same time you sensed the pain, your body's self-healing mechanism was turned on. A multistep signaling cascade involving a dozen different proteins in the blood started immediately after the cut, calling for platelet cells to clog arround the wound and form a plug to stop the bleeding. How does our body sense a wound so fast? A possible answer is that the platelet cells carry on their surface a sensitive molecular flow sensor, a protein called GP1b, which senses erroneous blood flow caused by a cut in the blood vessel. It has been hypothesized that a small segment located on the alpha-subunit of GP1b, called the ß-switch, transforms from a random coil to a ß-hairpin in the presence of shear flow (see movies). The ß-switch, in its ß-hairpin form, is able to bind to a protein called von Willerbrand factor, which will then anchor the platelet to the damage site. To test this hypothesis, researchers resorted to computer simulation using the program NAMD and mimicked blood flow around GP1b. As reported recently, the researchers discovered that a long suspected part of GP1b indeed changes shape under flow conditions, the likely trigger of the body's self-healing system. For more information see our molecular flow sensor web site.
image size:
117.5KB
made with VMD
Living cells are brimming with activity, much of it due to their
manifold molecular machines pulling cargo, importing and exporting
molecules, digesting food molecules and transforming their energy,
reading and copying genetic messages, or synthesizing proteins from
these messages (the latter done by the ribosome). Static structures of
the molecular machines have been resolved through crystallography:
machines pressed into the confinement of crystals and frozen into
inactivity reveal their atomic level geometry through this methodology.
However, many machines, for example the ribosome, undergo large
conformational transitions during their cyclic action, but active
motions are hard to view in atomic detail. A way out is offered by
electron microscopy which freeze-shocks machines into states
characteristic for action cycle intermediates. Unfortunately, the method
does not yield atomic resolution images, leaving the chemical
detail needed for a comprehension of the mechanisms blurred.
Computational methods can be used to bridge the resolution gap: atomic level
structures of non-functional states of the machines captured in crystals
are deformed to match electron microscopy images. Until recently, the method
worked well only for very small machines. Now a team of electron
microscopists and computational biologists using NAMD extended the
method to common size machines and reported
its successful application to the ribosome, providing astonishing detail
about ribosome dynamics and function. For more details, see our MDFF website.

image size:
32.4KB
made with VMD
Proteins carry out most functions in living cells, from import
of food substances to chemical synthesis to motion to signaling.
Proteins are chains of amino acids like GLSDGEWQLVLNVWGKVEAD... where
each letter stands for one of twenty amino acids that are the
building blocks of proteins, i.e., G for glycine or L for leucine.
In general, sequences of proteins native to cells fold into unique
three-dimensional structures capable of executing the proteins'
specific function. Living cells store the amino acid sequences
of their many different proteins in the form of DNA sequences,
safeguarding them in the cells genome. On demand, the DNA sequences
are translated according to the famous genetic code into amino acid
sequences. The amino acid chains of newly synthesized proteins
have to fold into the proper structure, an essential process
scrutinized by biologists for decades. The folding process often takes
milliseconds or longer, but recently proteins were identified
that fold within microseconds. This was still a time too long to be
simulated through molecular dynamics which could reveal folding in
atomic level detail. However, improvements of NAMD have now made
simulations of 10 microseconds possible and in a recent report
experimental and computational biologists describe a joint study
of a protein segment, known as the WW domain, over this timescale.
The great increase in simulation time revealed intricate details
of WW domain folding, but also points to a need to further improve
the computational model (force field) used to simulate
proteins. See also our protein folding web site.
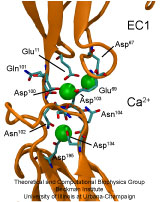
image size:
97.1KB
made with VMD
Adhesion between human cells organizes our body into its organs and
parts. The adhesion comes about through an intricate system of
molecules that perform their task in a highly selective manner such
that the body's different types of cells will find the right cells and stick to them.
This selectivity leads to tissue differentiation and the
organization of organs as complicated as the brain. Cadherin proteins
form a particular family of such adhesion molecules. Interestingly,
they glue cells together only in the presence of calcium. Some
members of the cadherin family of proteins are also involved in the
transduction of sound and cadherin mutants are known to cause
hereditary deafness (see the April 2005 highlight, "Hearing: Turning Sound into Voltage").
How cadherins selectively bind to each other
and the role of calcium was not well understood, but now molecular
dynamics simulations have offered magnificent insight into
calcium's role as recently reported.
The simulations took advantage of parallel supercomputers and
NAMD's ability to harness their
power. The simulations revealed that in the absence of calcium
cadherins stick out of cell surfaces like ends of loose rope; in the
presence of calcium cadherin molecules turn into stiff hooks that link
cells together. The calcium-induced links can withstand the strong mechanical forces
that arise between cells much larger than cadherin (more on
our cadherin website).
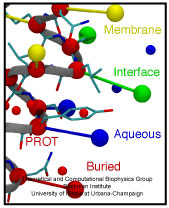
image size:
227.2KB
made with VMD
The environment of cells can undergo drastic changes, for example
from dry to wet, in which case cells shrivel or swell. However,
they are protected from bursting by a system of safety valves in
their cellular membranes that open and release cellular content.
Some of the valves open already at low membrane tension, but only
little, others open only at higher tension, but wide and without
filtering outflow. The mechanosensitive channel of small
conductance, MscS, is a low pressure safety valve in bacterial cells
(see the Feb
2007 highlight, "Observing and Modeling a Crucial Membrane Channel", the May 2006
highlight, "Electrical Safety Valve", and the Nov 2004
highlight, "Japanese Lantern Protein"). MscS is able
to rescue cells about to burst by releasing small solutes through a
large and transient opening in the cell membrane, thereby relieving
internal pressure. The only way to learn how MscS performs this vital
task is by determining its atomic-level structure under native
conditions. However, the only structure available for MscS was
obtained for the purified and crystallized protein; inspection of
the structure left doubt that it shows a functional protein, i.e., a
closed safety valve. Now a team of experimentalists and modelers
report
the structure of MscS seen in its natural membrane
environment. In their approach, simulations incorporate information
from so-called paramagnetic resonance measurements experiments. This
finding is yet another case where the combination of modeling and
observation offers entirely new close-up views of living cells (more
on our MscS website).
image size:
332.0KB
made with VMD
Bleeding through physical injury is stopped in animals through the formation of blood clots. Such clots, actually arising often in blood vessels without injury, can rupture due to the blood's shear forces and obstruct upstream smaller vessels leading to life threatening stroke, pulmonary embolism, and heart attack. Hence, a blood clot must be both mechanically stable to stop bleeding, yet elastic enough to avoid rupture. Fibrin, the main component of a blood clot, possesses the stated mechanical properties in healthy individuals, but in pathological circumstances needs to be managed through medication. Unfortunately, preventive treatment of blood clots is still a black art since the
molecular basis of fibrin elasticity is unknown. Clinicians at the Mayo Clinic teamed up with computational biologists at the University of Illinois to investigate this elasticity, focusing on the protein fibrinogen, the building block of fibrin. The clinical researchers stretched individual fibrinogen molecules measuring the force needed to extend the molecules. They found a characteristic force - extension relationship and its dependence on blood pH and calcium concentration, but they could not interpret the finding chemically, a prerequisite for improving blood clot chemical management. The clinical researchers joined forces with computational biologists who could reproduce the measured force - extension relationship in steered molecular dynamics using NAMD. The simulations starting from known crystallographic structures of fibrinogen offered a full, i.e., atom resolution, chemical picture of fibrinogen elasticity. As reported recently by the clinical and computational researchers the insight gained opens new avenues for blood clot treatment. For example, it was found that pH and calcium concentrations alter the stiffness of blood clots, thereby opening pharmacological avenues for controlling the incidence of pathological blood clots. More on this investigation can be found here.
Biological cells protect their interior through their cellular membranes, yet rely on import of nutrients. They have evolved for this import fast conduction channels that include reliable checkpoints distinguishing desirable and undesirable compounds. A checkpoint puts up a veritable obstacle course that only the right compounds can pass quickly. Understanding the channel design is difficult due to lack of detailed experimental data on nutrient dynamics. Presently, the most detailed information comes from viewing channel dynamics computationally, starting from static crystallographic structures. A recent study investigated how glycerols, small nutrient molecules needed by some bacteria, pass through checkpoints realized through the membrane protein aquaporin (see also highlights Gas Molecules Commute into Cell - Mar 2007, Aquaporin and the Cambridge Five - Oct 2006, Cellular Faucets - Feb 2006). Aquaporin furnishes four parallel channels that were monitored computationally using NAMD and a novel algorithm that explores the channel energetics quickly enough to be methodologicaly feasible on today's computers. The results show how the physical characteristics of glycerol, for example the molecule's ability to form so-called hydrogen bonds, its electrical dipole moments, its diffusive mobility and intrinsic flexibility are probed along the channel, discriminating glycerol from other molecules. More on computational investigations of aquaporin here.