Highlights of our Work
2026 | 2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
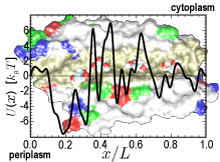
image size:
184.8KB
made with VMD
Atomic force microscopy measurements of the surfaces of cells lead to a bizarre discovery: single cells twitch with their surfaces. When the microscope feeds a microphone one hears the murmur, shrieking, or singing of cells. But why do cells sing? A recent report (covered in a news report) suggests that the twitching motion of cell membranes can help cells directionally transport nutriments across their cell wall. Earlier results of computer simulations that described the energetics of glycerol (a nutriment for E. coli) conduction across an aquaglyceroporin membrane channel (see June 2002 highlight) combined with a mathematical analysis of channel transport concludes that the twitching motion of cells leads the channel to act as a ratchet: the twitching moves the glycerol, the channel energetics discourages backslip such that the glycerol moves in one direction. The direction is outside - in when glycerol is in short supply, and inside - out when there is too much of it. The finding has deep implications for other biological channels showing how they can use energy from pulsating membranes to drive solutes through.
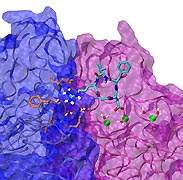
image size:
198.4KB
made with VMD
Biological cells must be capable of attaching themselves to their surroundings. For this purpose they utilize fibrillar proteins, such as fibronectins, that grasp cells through cell surface receptors integrins. The latter act as snap fasteners to the extra-cellular fibrils. The growth, movement, and survival of cells are all dependent on the ability of integrins to fasten cells upon intra-cellular signals or to signal inwards that something has become fastened on the cell surface. The major fastener on integrins are simple divalent ions like Mg++ or Ca++ that can adhere to specific molecules with amazing strength, even though the interaction at the cell surface is exposed to water. Computer simulations using NAMD, reported recently, revealed a dynamic picture of the interactions used by cells to link themselves to the extra-cellular matrix. They showed that it is actually a brave water molecule that is recruited by integrins as a protective shield for the interaction. The simulations provide for the first time a detailed view of how cell tissues are stabilized through surface ions against mechanical stress.
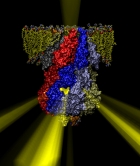
image size:
172.0KB
made with VMD
Many bacteria hang into their cellular membranes proteins that look like a Japanese lantern. These proteins have a flexible cylinder (pore) that crosses the membrane and opens and closes depending on membrane tension; from the cylinder hangs a balloon with seven small openings around its equator (see figure). The apparent function of the Japanese lantern protein, aptly called mechanosensitive channel of small conductance (MscS), is to protect the bacterial cell against osmotic stress: when a bacterium finds itself suddenly in an aqueous environment entering water can burst the cell. Before this happens the cell membrane experiences tension that opens the protein pore, permitting passage of water and ions, the efflux being controlled through the protein balloon. A recent study explores the dynamical properties of MscS, e.g., pore closure and opening as well as ion conduction, by means of molecular dynamics simulations using NAMD. Embedding the large protein into a lipid bilayer and water led to a simulation encompassing 220,000 atoms. Surprisingly, the protein balloon was found to control the arrangement of positive and negative ions through a peculiar pattern of charged, polar, and non-polar amino acids on its internal and external surfaces. This suggests that the Japanese lantern protein has a yet unknown second function in the bacterial cell.
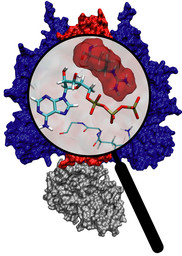
image size:
202.7KB
made with VMD
Molecular motors are efficient nanoscale machines destined to make any human designed engine look clumsy. F1-ATPase is such a machine - so powerful that a spoonful of it could produce as much torque as your car's engine. As part of the enzyme ATP synthase, the protein can work as an engine but also operate in reverse as a generator. In the latter mode it is responsible for the synthesis of the energy-rich molecule ATP that serves as fuel driving many processes in biological cells. It can also convert the energy stored in ATP into mechanical rotation. A recent study suggests that the analogy to a car's engine goes even further! A quantum chemical description of the reaction of ATP combined with a simulation of the protein revealed that an amino acid side group of the protein, called the "arginine finger", controls the progression of the catalytic event, much like a spark plug controls the combustion process in a car engine. The very extensive simulation made use of a powerful computer, the Jonas Cluster at the Pittsburgh Supercomputing Center. The investigation is yet another example for the important role of computational biology unraveling the secret behind the function of the machinery of living cells (more).
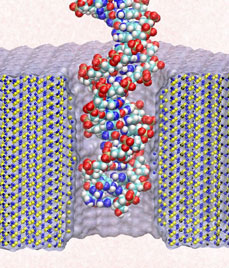
Electrical devices on computer chips built from silicon compounds have reached the small length scales of the building blocks in biomolecules, namely, the amino acids in proteins and the bases in DNA. Using beams of electron microscopes, electrical engineers drill nanometer wide pores into silicon wafers that contain a central layer only a few atoms thick. The engineers surround these pores with transistors and electrodes that can detect charges moving in the nanopore. Electrical fields across such synthetic nanopores can thread charged molecules like DNA through, and electrical signals stemming from single molecules transiting the pores can be recorded. Since the size of the nanopores compares with the dimension of DNA bases, the signals should eventually become precise enough to distinguish DNA bases, such that nanopores can become recording heads reading off sequences of DNA. While such ultrafast recording of DNA sequences is still a distant goal, the manufactured nanopores have been used already for sizing short strands of DNA as reported recently (report1, report2). Molecular dynamics simulations with NAMD and molecular graphics with VMD played a crucial role in imaging the dynamic events (movies available here) involved in recording single molecules of DNA and for optimizing the design of nanopores towards efficient threading and accurate recording. The landmark collaboration between computational biologists and device engineers promises to further unlock the great potential of biomedical nanotechnology.
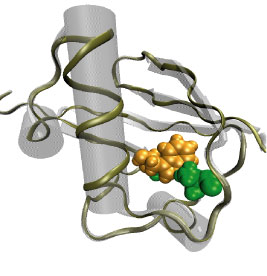
image size:
96.4KB
made with VMD
Proteins perform the many functions of biological cells. This ability arises from the particular three-dimensional structure into which proteins fold at physiological temperature. The quick and precise folding of proteins depends on their molecular environment, e.g., water or lipid membrane, and is being investigated in many laboratories today. A new study from a computational - experimental collaboration investigated the folding and resulting structure of a protein, ubiquitin, in ethylene-glycol, commonly known as antifreeze, mixed with water. In this mixture the folding could be monitored at very low temperature in "slow motion" and resolved in great detail. Computational modeling using NAMD and VMD suggested that adding antifreeze to water leaves ubiquitin folding unaffected and this was born out indeed by further observation. Antifreeze is offering now a wider window into the study of the amazing abilities of other proteins.
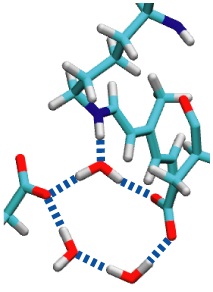
image size:
79.3KB
made with VMD
Sun light offers abundant energy fueling life on earth. Primary users of this energy are photosynthetic life forms. Remarkable in this regard are certain halophilic bacteria that developed a protein, bacteriorhodopsin (bR), which acting as a light driven proton pump turns sun light into a voltage gradient across the cell wall. Each photon absorbed primes bR to transfer of a proton. The bR pump had been discovered 30 years ago, yet the mechanism of the pump is still basically unknown. A key step towards establishing the mechanism would be identification of the initial energy form stored after light absorption. While some researchers suggest energy is stored highly localized in bR, others recognized that given the soft nature of proteins storage should be spread over many degrees of freedom. Widely accepted candidates are torsions of retinal, the linear molecule in bR that actually absorbs the light. A collaboration between computational and experimental groups carrying out unprecedented calculations and spectroscopic observations of water inside bR has now reported that a key fraction of light energy is stored in a geometrically distorted so-called hydrogen bond network involving retinal, three water molecules, and three amino acid side groups of bR. The results demonstrate dramatically that, in oder to reveal mechanisms underlying protein function, structural details at the sub-Angstrom level need to be resolved; only computational modeling guided by observation is presently capable of such resolution.
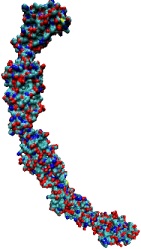
image size:
170.9KB
made with VMD
One of life's great achievements is the development and maintenance of multi-cellular organisms, from an embryo to adulthood. Multitudes of cells need to be sorted and resorted into tissues, organs, and body of living beings. One strategy towards this end is to endow cells with so-called adhesion proteins that connect a mechanical framework inside cells through the cell membrane with other cells. A key type of adhesion protein is cadherin (calcium-dependent adherent protein) that stretches through the cell surface five-tandem domains. The outermost domain can stick to a cadherin molecule from an adjacent cell. Crystallography provided the molecular structures of cadherin pairs and resolved in atomic detail the cadherin-cadherin contact between cells. This prompted a collaboration that aimed at probing the adhesion strength of cadherin pairs through steered molecular dynamics simulations stretching the pairs apart. Results of the simulations were reported in a recent publication that employed NAMD as well as VMD. As shown by crystallography, the cadherins each insert a tryptophan residue into the other protein. The link thereby established can be broken only through strong forces that induce a step-wise slippage of the residues first out of their binding pockets and then along the protein surface. This scenario suggests a mechanism for selectivity among cadherins, i.e., why among the various cadherins found on the surfaces of cells some adhere much better to each other than others.
Since Leuwenhook, microscopic images of living matter have been produced with radiation, from light to X-rays. With the advent of ever more reliable computational methodologies, molecular dynamics has reached the status of a trusted research instrument. This instrument is particularly powerful for imaging nanoscale, i.e., 10-100 nanometer size, systems. This month our group brought three computers on-line that can serve to image nanoscale systems, three 48-processor rack-mount Xeon clusters (pictured) running our MD program NAMD. One such cluster, a nanoscale system of 300,000 atoms imaged over a nanosecond at the most advanced simulation conditions possible today, requires four days of computing. The Clustermatic software from Los Alamos National Laboratory makes each cluster of 48 processors appear to biomedical researchers as a single machine and allows interactive simulations to temporarily displace long-running NAMD jobs. The clusters have been used already for a study of balance and hearing in the human inner ear. These senses are intrinsically mechanical, relying on hair cells to convert vibration to ion channel modulation. Ankyrin, a protein formed by repeats of a 33-amino-acid domain, is thought to act as a molecular spring in mechanotransduction channels. Explaining the mechanism of ankyrin elasticity requires large simulations of 340,000 atoms that apply repeatedly stretching forces to the protein and monitor its response, revealing a mechanical behavior ideally suited for its biological function. The clusters are presently also used to design artificial nanopores for sequencing of DNA.
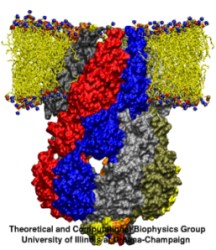
image size:
130.0KB
made with VMD
Computational and experimental biologists investigate jointly the physical mechanisms underlying the function of the molecular machines in cells. Simulations used to encompass biomolecular systems of 10,000 atoms, but recently the size increased tenfold. For example, simulations of aquaporins, a water channel, published during 2001-2003 (1, 2, 3) involved about 100,000 atoms and had been cited in connection with last year's chemistry Nobel prize; simulations of cadherin, a cell adhesion protein, and of ATPase, a key metabolic protein complex, published 2004 included a similar number of atoms. Simulations of over 200,000 atoms for a protein, lac repressor, that regulates genes, and for a protein, MscS, that is a mechanically gated membrane channel will be published later this year. Simulations involving over 300,000 atoms, on a protein, ankyrin, acting as an elastic spring in hearing, have been completed. The increase in the size of simulated systems is prompted by a revolutionary advance in crystallography that resolves ever larger structures of biomolecules and simulations are made possible through the great increase of computer resources at the NSF centers (PSC, NCSA). This marks the beginning of a new era in which systems like virus capsids and the ribosome, entailing 1-3 million atoms, will be studied, too. NAMD (see Highlight Dec 2002) is ready for the challenge posed by simulations needing 250-500 processors today and 1000-10000 processors in the future, to keep up with the developments in the biology laboratories (more).

image size:
130.0KB
made with VMD
Far from playing only the role of bricks and mortar as a mere divider between the inside and outside of cells or between parts of the cell, lipid bilayers are an active, tightly regulated cellular component whose physical properties are critical for the proper function of the membrane proteins contained within them. Lipid bilayers are the preeminent domain of computational biology since despite their considerable stability and impenetrability they form disordered films that are best described through computer modeling, albeit tested by observation. One of the largest molecular modeling projects achieved so far has been recently reported that employed NAMD to investigate the mechanical properties of cellular membranes. The systems simulated were made of lipids and water, composed of about 40,000 atoms, and simulated for over 100 nanoseconds. The simulations revealed that membranes, in terms of their mechanical properties, are far from being homogenous films; rather, they exhibit a delicate multi-lamellar structure of layers that alternatively tend to shrink and expand the membrane, inducing strong forces on all proteins and molecules entering. The lamellar character of the cell's membranes plays a key role for cellular processes such as osmotic regulation and may explain even the action of anaesthetics.
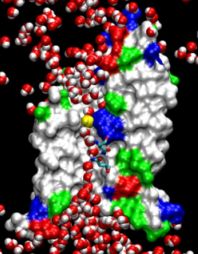
image size:
128.6KB
made with VMD
Biological cells are batteries charged by a voltage across their cell membrane. The voltage is maintained through a gradient of protons; the latter push back into the cell driving key cellular processes, e.g., the synthesis of ATP (see March 2004 highlight). But leakage of protons back into the cell is lethal. This generates a problem for water channels that exist in cell walls, e.g., in human tissues and plants (see October 2003 highlight), and conduct each about 10 million water molecules a second. Since protons are found in physiological solutions at a fraction of 1/10 million each channel should pass along with the water a proton per second, draining the cellular battery. The cells' water channels amazingly prevent this leakage. The underlying mechanism was suspected to be connected with an orientation pattern of water in the channel enforced by an internal electrostatic field (see May 2002 highlight) that neither allows protons to hop through the channel, overtaking the water transport speedwise, nor permits protons to flow along with the water. Now two collaborative studies (1, 2) have confirmed the suggestion, finding an energy barrier high enough to reduce the chance of proton passage to less than a proton per day. (more)
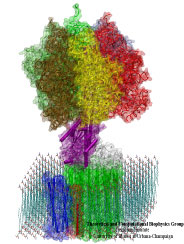
image size:
245.6KB
made with VMD
What will car motors look like in a million years? It's hard to tell, but biological cells seem to have found an ideal engine that they use since the early days of evolution. A spoonful of their engines generates about as much total torque as the strongest car engine today. The engine is called FoF1-ATP synthase and synthesizes the molecule ATP by combining two generator-like motors, Fo-ATPase and F1-ATPase, coupled through a single axle, one motor (Fo) that converts the cell's electrical energy into rotation, another one (F1) that converts rotation into chemical synthesis (see November 2003 highlight). ATP synthase, found throughout the whole kingdom of life, can also work in reverse, turning ATP into electrical energy. Cells typically use the energy of sun light or of food to generate an electrical potential by pumping protons that carry a positive charge across their cell membrane. The energy stored drives the protons back through Fo-ATPase enforcing rotation of the axle; the rotation in turn induces ATP synthesis in F1-ATPase. In one of the largest computational and mathematical biomodeling projects undertaken to this day and reported recently, researchers build from available disjoint structural data a model of Fo-ATPase and demonstrated its function as a motor turning proton conduction into rotation of a cylindrical protein complex. By linking nanosecond molecular modeling to a mathematical model of the motor's key elements, they could follow Fo-ATPase function properly, even when the load arising from driving synthesis in F1-ATPase was added. FoF1-ATP synthase being one of the largest molecular machines in biological cells, modelers needed to employ for its study the most advanced tools, NAMD and massively parallel computers, along with a new approach that combined molecular dynamics and stochastic mathematics.
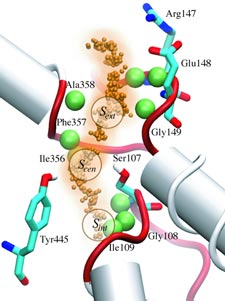
image size:
90.4KB
made with VMD
When their environment gets high in hydrochloric acid, bacteria must protect themselves from chloride ions leaking into the cells. Some bacteria rely on so-called ClC chloride channels to quickly evacuate these ions without letting other small particles pass through. A recent paper investigates the mechanism of chloride transport in a ClC channel from the bacterium E. coli. By means of computer simulations using NAMD, researchers visualized the pathway taken by chloride ions as they pass through the channel and identified how the channel's protein architecture optimizes ion conduction. The relevant architectural features have also been observed in aquaporin water channels (see November 2003 highlight) and the potassium ion channel, but are rarely seen in other proteins. The new discovery demonstrates how much computer simulations are contributing to the emerging picture of life's membrane channel design that is critical for functions of biological cells ranging from the maintenance of our body's hydration to electrical signaling in the brain.
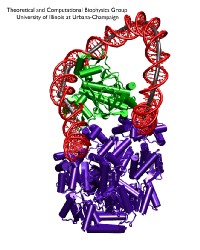
image size:
144.6KB
made with VMD
Bacteria must respond quickly to food sources in their environment. A common source of food for the bacterium E. coli is lactose; the bacterium's DNA contains genes for proteins needed for import and metabolic degradation of lactose and related sugar molecules. When lactose is lacking, a regulatory protein called lac repressor prevents the needless expression of lactose processing proteins; in doing so, lac repressor is assisted by a second protein, called catabolic activator protein (CAP). To understand the roles of the two proteins in regulating DNA, a recent report employed an advanced mathematical model for the flexibility of DNA and showed how lac repressor and CAP force DNA to form a loop such that the local genetic information cannot be expressed; the loop is held at its ends by the lac repressor and CAP inserts itself into the loop, adjusting and stabilizing it. Together, lac repressor and CAP form a genetic switch, in fact, the first such switch discovered long ago and explained now in detail through mathematics. When lactose appears around the bacterium, the lac repressor becomes deactivated by the molecule and looses its grip on the DNA. In this case the DNA loop opens and the CAP present assists in the expression of the genes needed for lactose processing.