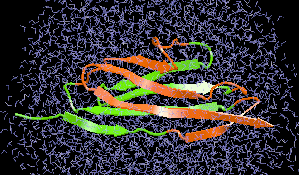Research Topics - Mechanobiology
Many cellular processes involve transformations and sustenance of mechanical forces. Forces arise as input, output and signals in cellular machines, such as ATP synthase, motor proteins, and cell surface receptors. Proteins functionally dealing with mechanical forces belong to the research subjects of the emerging new field mechanobiology. Examples among them are the giant muscle protein titin sustaining forces developed in muscle cells, the extracellular matrix fibronectin forming elastic fibrils connecting cells, mechanosensitive channels being switched on by tension applied to the cell membrane, integrins conveying mechanical signals across membrane, ankyrin amplifying weak force signals for the mechanotransduction process in hearing cells, lac repressor grappling with the regulated DNA, Fo-ATPase converting cell's electrical energy into rotation, and various ligand-receptor complexes. What is the physical mechanism underlying the mechanical functions of these proteins? Over the past decade the group has developed and applied a novel computational approach termed steered molecular dynamics to study the structure-function relationship of these mechanical proteins.
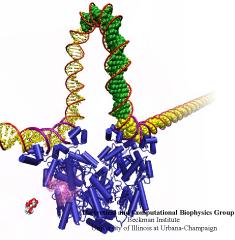
image size:
80.0KB
made with VMD
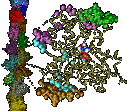
When Escherichia coli bacteria enjoy lactose and related food molecules in their environment, the cells quickly furnish proteins needed for import and metabolic digestion of the food. A set of genes, called the lac operon, is transcribed into messenger RNA that directs the synthesis of these proteins. When lactose is not available, the protein synthesis would be wasteful and, indeed, is prevented by locking the lac operon. This is achieved by a protein called lac repressor that forces the segment of the lac operon needed to initiate transcription into a loop, but that can be unlocked by a lactose molecule binding to the protein as soon as the food becomes available again. A recent study of the lac repressor combines a 314,000-atom protein simulation using NAMD with a multiscale simulation technique coupling the protein to the DNA loop. The calculations reveal how the lac repressor stretches out two "hands" grabbing the genomic DNA and then keeps a tight grip on the DNA wrestling it into a loop. The discovery is described on our website as well as in a popular article.
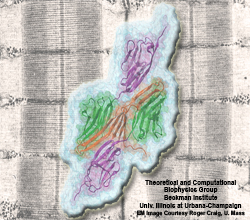
image size:
301.5KB
made with VMD
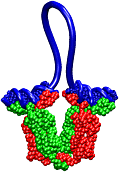
Muscle fibers are not rigid structures, but rather, they can both contract and extend in response to physiological demand. As a result, muscle sarcomeres must have a protective mechanism to prevent tearing and damage from overstretching. The giant protein titin fulfills this role by acting as a molecular rubber band, providing a passive resistance force during extension to restore the muscle fiber to its resting length. Conceivably, this rubber band must be anchored to a rigid structure in order to function. Biochemical investigations have speculated that the protein telethonin, located at the sarcomeric Z-disc, may serve this purpose. Genetic diseases related to defects in telethonin have been correlated with dilated cardiomyopathy and a form of muscular dystrophy. To date there have been no studies to determine how strongly bound titin is to telethonin. To explore this issue, we performed molecular dynamics simulations in order to test the strength of the newly resolved titin Z1Z2-telethonin complex. Our results, which have recently been reported (paper), reveal that the force required to dissociate titin from telethonin is significantly higher than that required to unfold isolated titin Ig-domains. This suggests strongly that telethonin is in fact an essential component of the Z-disc titin anchor. In addition, we find that telethonin anchors not just one, but two separate titin molecules, serving as a sort of molecular glue joining both titin molecules together through β-strand crosslinking (a structural motif also seen in fibril pathologies such as Alzheimer's, Parkinson, and Huntington's disease). Thus our simulations reveal also a fundamental architectural element of living cells, namely how cells glues their components together yielding strong mechanical connections. For more information on teletonin and the implications of our findings, see the following webpage here.

image size:
102.0KB
made with VMD
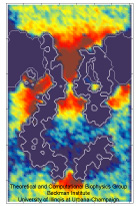
The ear is a sensitive and robust device, able to perceive the faint sound of flowing water and the thunderous blast of an air plane. Like a microphone, the ear transforms a complex, mechanical stimulus (sound), into an electrical signal, a voltage change in a nerve cell, that can be understood by our brain. This transformation is called "mechanotransduction" and is accomplished by a series of amazingly minute devices that each connect a soft spring to an ion channel, both located in specialized sensory cells, the hair cells of the inner ear. The springs, through their vibrations agitated by particular sound frequencies, control ion currents passing through the channels, thereby, modifying the hair cell internal electrical potential. This leads to neural signaling to the acoustic cortex of the brain. Recently reported molecular dynamics simulations using NAMD, some of the most extensive simulations accomplished to date both in size and duration, showed that the mechanical characteristics of hair cell signaling can be traced to a single protein, ankyrin, that acts as a helical spring. Imagine a soft spring that is extended several inches by the weight of a feather! Ankyrin is such a spring, but a billion times finer.
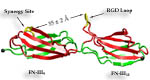
image size:
102.0KB
made with VMD
Cells constitute only part of animal tissue, much of tissue is made of the extracellular matrix (ECM), a flexible mesh composed of several classes of macromolecule. Fibronectin (FN) is a component of the ECM that acts as a specific adhesive, forming networks that connect cells to the giant fibrous molecules that make up the majority of the ECM. Proper FN fibril formation is required to maintain prop er cell migration, thus FN plays roles in diseases affecting growth, development, tumors , and wound healing. We have used SMD simulations to examine how the mechanical force s present in the ECM affect FN fibril formation.
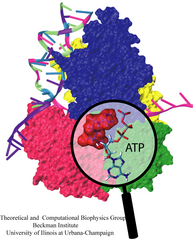
Actin protein units form filaments in the cells in order to manage the cellular shape changes, cell locomotion and chemotactic migration. The mechanical properties of the filaments substantially depend on the ATP hydrolysis by actin. Using molecular dynamics, we studied the ATP hydrolysis and the subsequent release of the inorganic phosphate.
A coarse-grained method of DNA modeling has been developed as a stepping-stone towards a multi-resolution approach to biomolecular modeling. The DNA structure is obtained after numerically solving a Kirchhoff system of equations, augmented with electrostatic and steric repulsion force terms. The method has been applied to restore the missing structure of a DNA loop clamped by the lac repressor.
The giant muscle protein titin, also known as connectin, is a roughly 30,000 amino acid long filament which plays a number of important roles in muscle contraction and elasticity. To examine in atomic detail the dynamics and structure-function relationships of this behavior, SMD simulations of force-induced titin Ig domain unfolding were performed.
The discovery of the crystal structure of the kinesin motor domain has made it possible to study kinesin's dynamics by computer simulations. To model pH- and nucleotide-dependent changes in the kinesin structure, we carried out conformational searches by simulated annealing. The conformational differences of the ATP-bound protein relative to the ADP-bound state can be attributed to a force-producing power stroke.
image size:
299.9KB
made with VMD
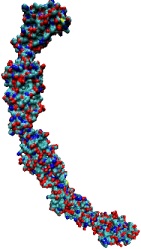
image size:
170.9KB
made with VMD
image size:
332.0KB
made with VMD
Bleeding through physical injury is stopped in animals through the formation of blood clots. Such clots, actually arising often in blood vessels without injury, can rupture due to the blood's shear forces and obstruct upstream smaller vessels leading to life threatening stroke, pulmonary embolism, and heart attack. Hence, a blood clot must be both mechanically stable to stop bleeding, yet elastic enough to avoid rupture. Fibrin, the main component of a blood clot, possesses the stated mechanical properties in healthy individuals, but in pathological circumstances needs to be managed through medication. Unfortunately, preventive treatment of blood clots is still a black art since the molecular basis of fibrin elasticity is unknown. Clinicians at the Mayo Clinic teamed up with computational biologists at the University of Illinois to investigate this elasticity, focusing on the protein fibrinogen, the building block of fibrin. The clinical researchers stretched individual fibrinogen molecules measuring the force needed to extend the molecules. They found a characteristic force - extension relationship and its dependence on blood pH and calcium concentration, but they could not interpret the finding chemically, a prerequisite for improving blood clot chemical management. The clinical researchers joined forces with computational biologists who could reproduce the measured force - extension relationship in steered molecular dynamics using NAMD. The simulations starting from known crystallographic structures of fibrinogen offered a full, i.e., atom resolution, chemical picture of fibrinogen elasticity. As reported recently by the clinical and computational researchers the insight gained opens new avenues for blood clot treatment. For example, it was found that pH and calcium concentrations alter the stiffness of blood clots, thereby opening pharmacological avenues for controlling the incidence of pathological blood clots. More on this investigation can be found here.
Bacterial cells can swim and use for this purpose one or more flagella, whiplike appendages that exceed the length of the cell severalfold. The flagella are made of many thousand copies of a protein called flagellin, arranged in a helical fashion such that the flagella are hollow inside, forming a very long channel. When the flagella are rotated by the cell counter-clockwise, the cell swims straight; when they are rotated clockwise, the cell turns to a new direction. Through swimming and turning the cell searches its habitat for food and avoids trouble. But sometimes a flagellum breaks and needs to grow back. At this point starts an amazing process: the cell makes new flagellin and pumps the unfolded protein into the flagellar channel, extending its length. This is like squeezing toothpaste out of a tube, except in reverse, like pumping toothpaste into the tube at the toothpaste factory, and the tube is extremely long. Now researchers have described the process that makes flagella grow step-by-step through a combination of mathematics, physics, and molecular modeling using NAMD. As reported, the researchers reproduce the time course of growth as well as the length of the growth and also explain how friction of the protein paste is kept extremely low to make the flagella grow many times the length of the cell itself. More information here.
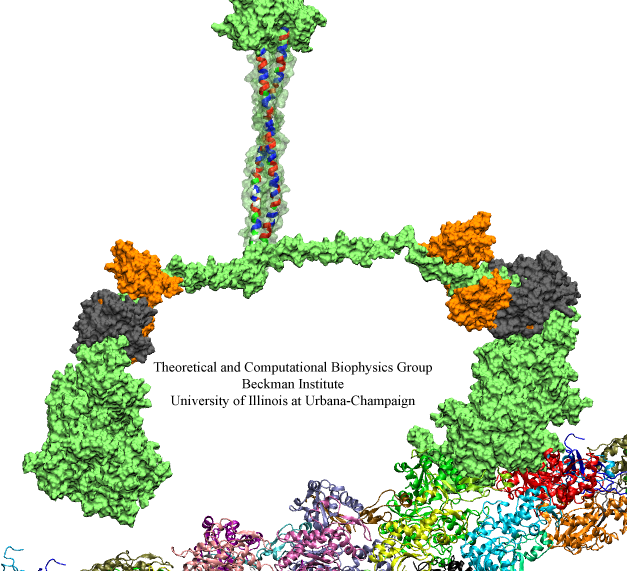
image size:
86.3KB
made with VMD
Cells are often compared to busy metropolitan cities, and in this analogy, motor proteins are the delivery trucks running on the roadways and transporting goods to different part of the cell. Motor proteins are found in almost all eukaryotic cells, and they convert chemical energy using ATP hydrolysis into mechanical work that powers their movements along cytoskeletal tracks. Three classes of motor protein superfamily have been characterized: myosin, kinesin, and dynein. While kinesin and dynein move along the microtubule network, myosin moves along actin filaments. Over the years, the structural basis of how motor proteins work has been established by solving the x-ray and cryo-EM structure for different domains of several motor proteins. What remains missing is the dynamic process connecting different states of a motor protein's catalytic cycle. By teaming up with experts of single-molecule techniques from the Selvin lab here at UIUC, we hope to gain a molecular understanding of the stepping mechanism of motor proteins.
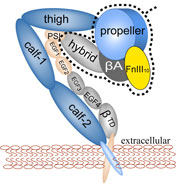
The ability of striated muscle to stretch and relax, in an elastic manner, is crucial for its role as a force bearing component in higher organisms. An individual muscular contractile unit, called the muscle sarcomere, extends beyond its equilibrium length immediately following contraction, and a passive restoring force arises from a protein called titin, whose elastic response restores a muscle fiber to its resting length. Titin is the largest known protein, with a length greater than 1 µm and a molecular weight of up to 4 MDa. It is built from a modular construct of ~300 tandem repeats of predominantly immunoglobulin-like (Ig) and fibronectin type III (FnIII) domains, and from flexible random coil-like PEVK (rich in proline, glutamate, valine, and lysine) regions; there exist furthermore single Ig-domain insertions that distinguish isoforms of titin in cardiac and skeletal muscle.