Highlights of our Work
2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
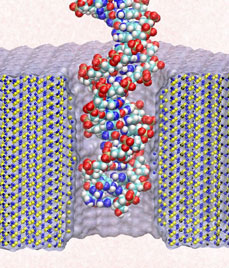
Electrical devices on computer chips built from silicon compounds have reached the small length scales of the building blocks in biomolecules, namely, the amino acids in proteins and the bases in DNA. Using beams of electron microscopes, electrical engineers drill nanometer wide pores into silicon wafers that contain a central layer only a few atoms thick. The engineers surround these pores with transistors and electrodes that can detect charges moving in the nanopore. Electrical fields across such synthetic nanopores can thread charged molecules like DNA through, and electrical signals stemming from single molecules transiting the pores can be recorded. Since the size of the nanopores compares with the dimension of DNA bases, the signals should eventually become precise enough to distinguish DNA bases, such that nanopores can become recording heads reading off sequences of DNA. While such ultrafast recording of DNA sequences is still a distant goal, the manufactured nanopores have been used already for sizing short strands of DNA as reported recently (report1, report2). Molecular dynamics simulations with NAMD and molecular graphics with VMD played a crucial role in imaging the dynamic events (movies available here) involved in recording single molecules of DNA and for optimizing the design of nanopores towards efficient threading and accurate recording. The landmark collaboration between computational biologists and device engineers promises to further unlock the great potential of biomedical nanotechnology.