VMD-L Mailing List
From: Akash Banerjee (akashneon_at_gmail.com)
Date: Mon Feb 01 2021 - 11:10:34 CST
- Next message: Nathan Kern: "Re: how to search for existing topology"
- Previous message: Vermaas, Josh: "Re: how to search for existing topology"
- In reply to: Gumbart, JC: "Re: Re: Unable to fit dihedral to QM target data in FFTK"
- Next in thread: Daniel Fellner: "Re: Re: Unable to fit dihedral to QM target data in FFTK"
- Reply: Daniel Fellner: "Re: Re: Unable to fit dihedral to QM target data in FFTK"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]
Hello,
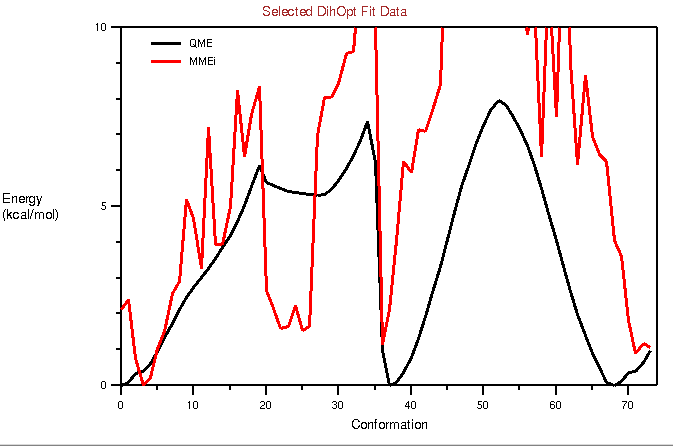
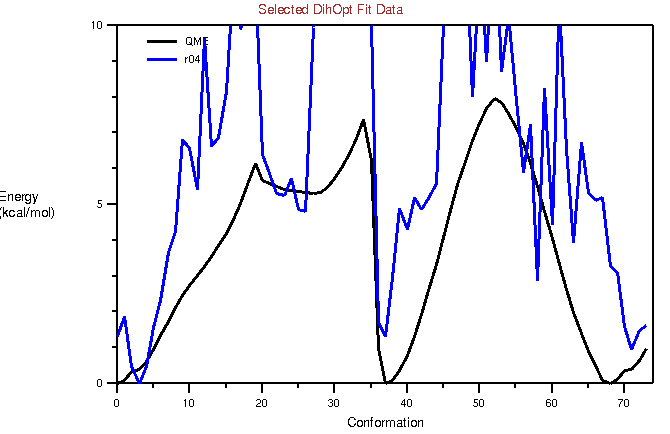
I have attached the potential fits before and after refinement. I am seeing
2 peaks, and hence I tried refitting with a periodicity of 2. However, that
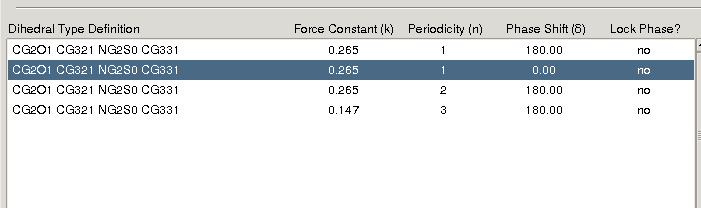
does not really reduce the RMSE. When I assign 2 more dihedrals with a
periodicity of 1, there is minor improvement in the RMSE (3.4->2.2). Since
I am adding so many dihedrals (4 in number, please see
attached dihedrals.png), I guess I am over fitting.
More details about my system: I have a polymer backbone with aromatic side
chain groups. I just need to use FFTK to parameterize the dihedral that
connects the aromatic side chain to the backbone.
>From my side, I am thinking that these could be potential issues:
1. Initial geometry (fixed it with geometry optimization in FFTK itself)
2. How I am sampling the dihedral (just following the tutorial, i.e do a
scan from -180 to 0, and then 0 to 180)
3. Sampling rate ( at intervals of 5 degrees to get good statistics)
4. How I upload the QM target data to FFTK (first negative scan, and then
the positive scan)
Please help me figure out what could be wrong. Since I am new to QM and
FFTK, I could be missing some fundamental details.
Thank you.
Kind Regards,
Akash
On Mon, Feb 1, 2021 at 10:49 AM Gumbart, JC <gumbart_at_physics.gatech.edu>
wrote:
> I think it should be possible to fit a single dihedral perfectly. Does the
> potential look unusual in some way?
>
> Best,
> JC
>
> On Jan 28, 2021, at 5:28 PM, Akash Banerjee <akashneon_at_gmail.com> wrote:
>
> Hi VMD Developers,
>
> I just wanted to follow up on the above ticket. Should I provide my input
> files to clarify the issue? Please let me know if I could provide any
> additional information that could help in this regard.
>
> Thank you.
>
> Kind regards,
> Akash
>
> On Mon, Jan 18, 2021 at 5:31 PM Akash Banerjee <akashneon_at_gmail.com>
> wrote:
>
>> Hi FFTK/VMD developers,
>>
>> I am trying to resolve a dihedral in a polymer chain. I have generated
>> the QM target data, but I am facing difficulties in fitting the dihedral to
>> the target. I have enlisted my protocol below. Please have a look and
>> advise me on how I could correct it.
>>
>> 1. Generated an optimized trajectory (fit to QM data), and used the
>> modified PDB file for further studies.
>> 2. Scanned the dihedral of interest (my molecule has several dihedrals).
>> I only scan the one that is associated with a high penalty factor (assigned
>> by CGenFF). I followed the routine given in the tutorial, i.e generated a
>> QM input file for a negative (-180 to 0) and a positive (0 to 180) scan. I
>> kept an interval of 5 degrees for both scans for better results.
>> 3. The QM simulations were run on a remote resource, and QM log files
>> were successfully generated.
>> 3. In the optimize dihedrals tab, I uploaded the modified PDB , PSF, PAR
>> and 2 QM target log files. I selected the dihedral that requires
>> optimization, and I clicked the "run optimization" button. The resultant
>> RMSD is 3.54.
>> 4. I do refinements by adding dihedrals with different periodicity and
>> phase shift angles. I referred to table 19 in this link:
>> https://www.ks.uiuc.edu/Training/Tutorials/science/forcefield-tutorial/forcefield.pdf
>> <https://urldefense.com/v3/__https://nam12.safelinks.protection.outlook.com/?url=https*3A*2F*2Fwww.ks.uiuc.edu*2FTraining*2FTutorials*2Fscience*2Fforcefield-tutorial*2Fforcefield.pdf&data=04*7C01*7Cgumbart*40physics.gatech.edu*7C1fcaa9cfec7448969c2808d8c3e11e5f*7C482198bbae7b4b258b7a6d7f32faa083*7C0*7C0*7C637474718940642800*7CUnknown*7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0*3D*7C3000&sdata=cuzbs3vIOUSGSS3uZ*2BV*2BPJPHpZhrEmRT57cs9FshN*2F0*3D&reserved=0__;JSUlJSUlJSUlJSUlJSUlJSUlJSUlJSUl!!DZ3fjg!ulm7u_TLt0cJwwNON_1UNu7kqzzOu3YjJsKdBgHDcLfm2I1Y5iy3F4JF8-akBEoF8w$ >
>>
>> After trying several combinations (i.e all possible periodicity, phase
>> shift angles, optimization algorithms and tolerance values), I get a RMSD
>> of 2.2. I am not able to reduce the RMSD beyond the 2.2 mark.
>>
>> Do you have any insights into how I could get lower RMSD values, in turn
>> improving the dihedral fitting protocol?
>>
>> Thank you.
>>
>> Kind Regards,
>> Akash
>>
>>
>>
>
> --
> akash
>
>
>
-- akash
- Next message: Nathan Kern: "Re: how to search for existing topology"
- Previous message: Vermaas, Josh: "Re: how to search for existing topology"
- In reply to: Gumbart, JC: "Re: Re: Unable to fit dihedral to QM target data in FFTK"
- Next in thread: Daniel Fellner: "Re: Re: Unable to fit dihedral to QM target data in FFTK"
- Reply: Daniel Fellner: "Re: Re: Unable to fit dihedral to QM target data in FFTK"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]



