Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
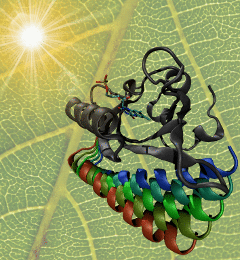
image size:
1.9MB
made with VMD
The ability to sense light is crucial for both plants and animals; animals use their vision to navigate and interact with their surroundings, whereas plants grow toward light to optimize photosynthesis. One of the most important photosensors in plants relies on tiny molecular switches known as LOV domains. When light strikes a LOV domain, it causes the formation of a single chemical bond; the unique structure of the LOV domain converts this subtle change into a protein unfolding event that triggers signaling. The mechanism through which LOV domains amplify bond formation into large-scale molecular motion is of great interest both for designing light-activated proteins for synthetic biology applications, and as a model for understanding the harder-to-study molecular switches that govern most of the functions of living cells. As reported recently, researchers used a series of long-timescale molecular dynamics simulations to show the locations of molecular levers that allow light-induced bond formation to rearrange the entire structure of the LOV domain. The simulations highlighted two main paths of information flow from the heart of the photoreceptor to the surface of the protein, giving unprecedented insight into the function of this light-activated molecular switch. More information is available on the biological photoreceptors website.



