Highlights of our Work
2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
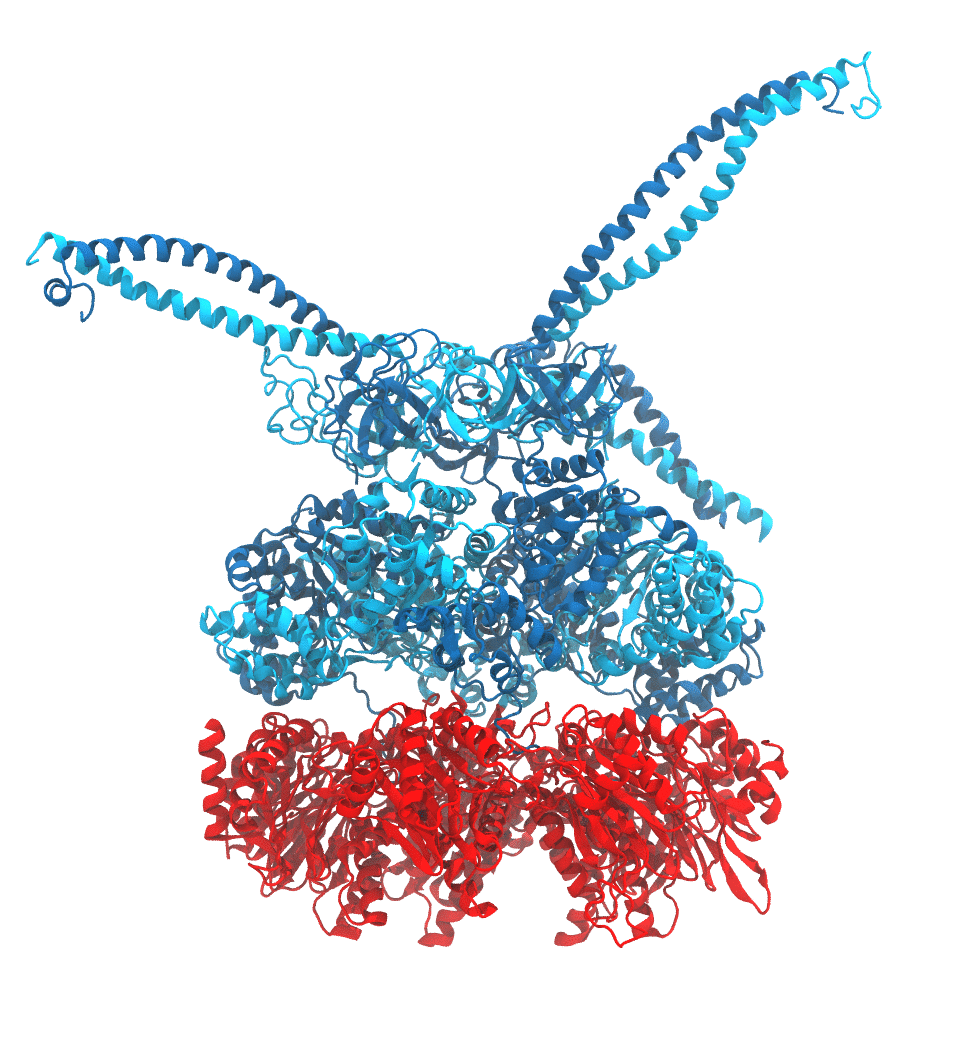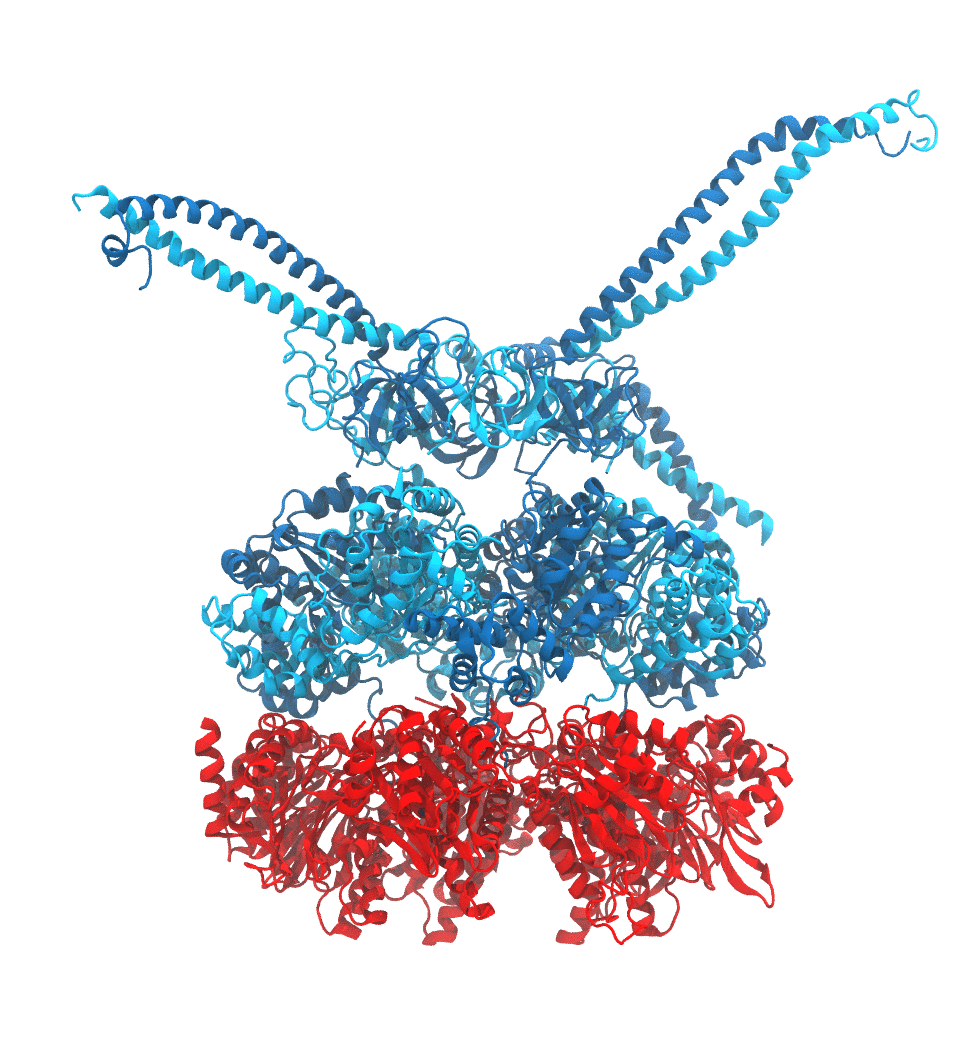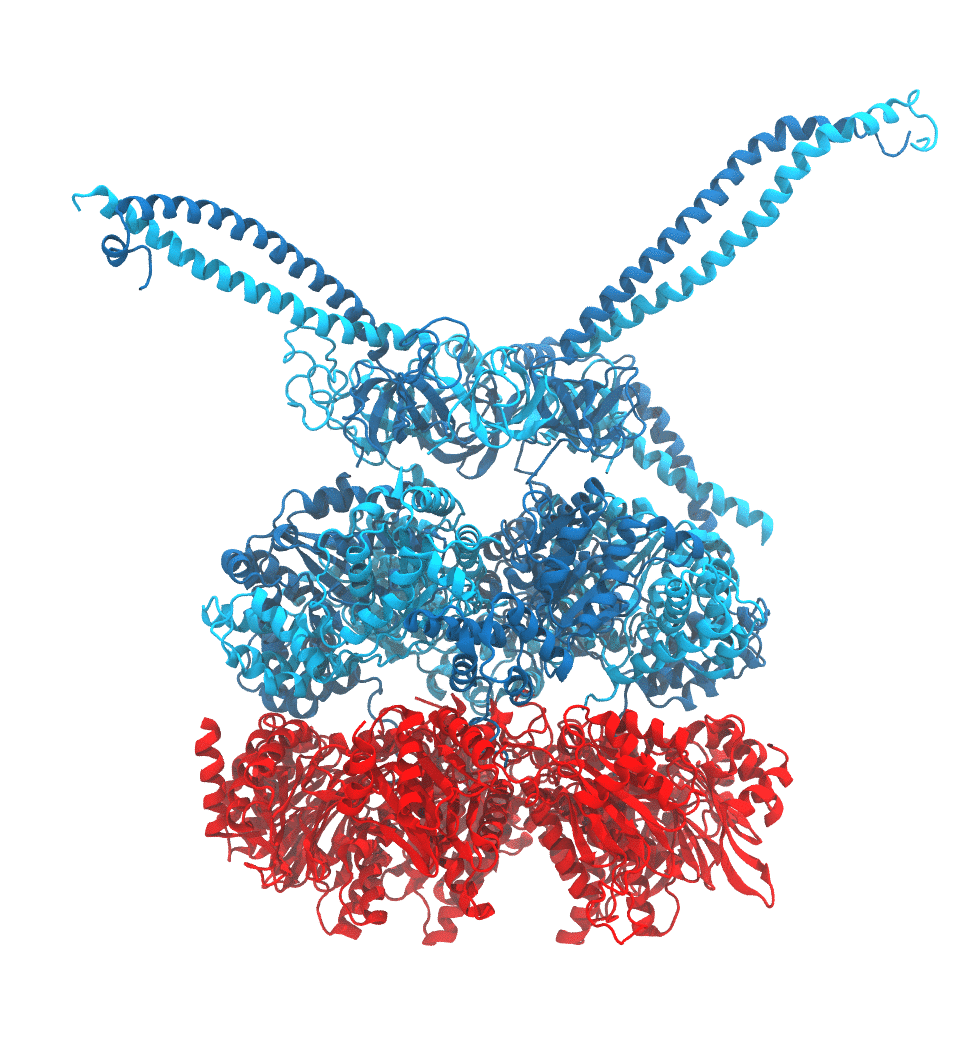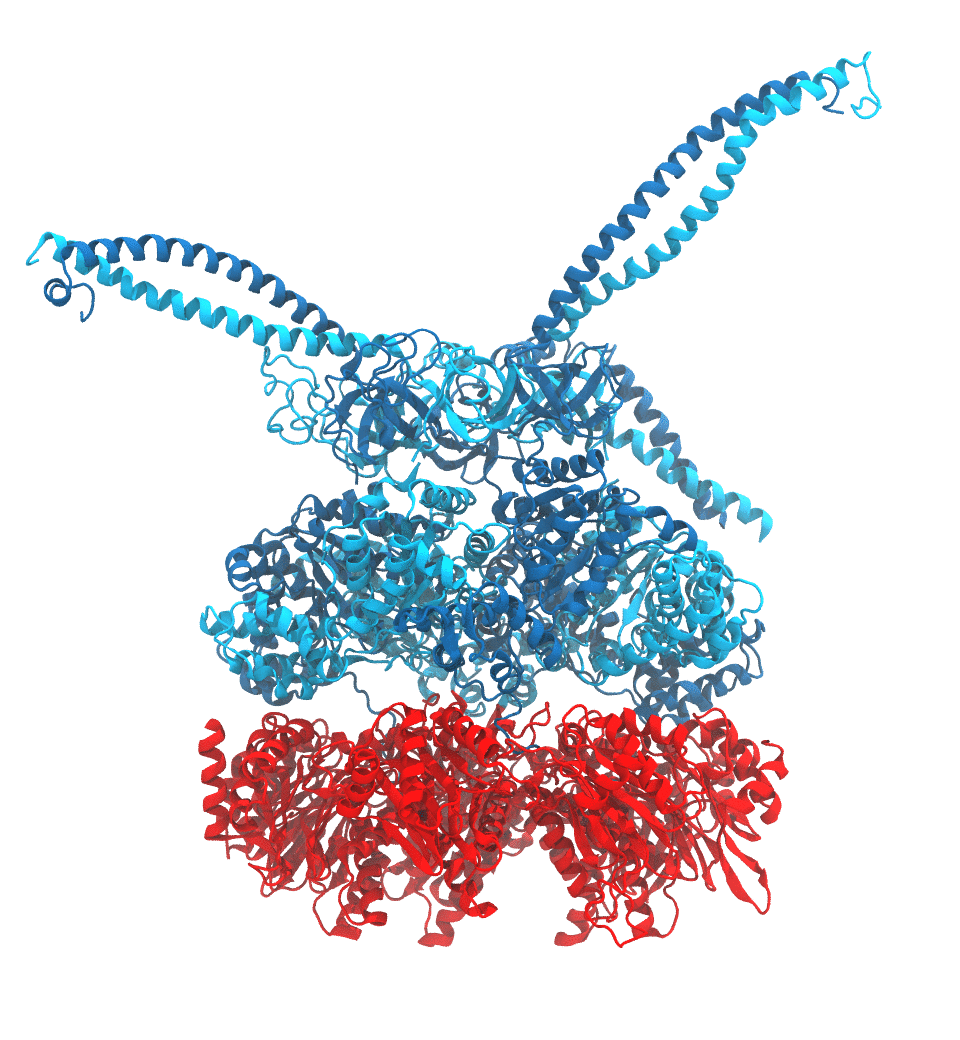
Protein recycling is a key process crucial to a wide spectrum of regulatory processes
within living cells. The executive player in this process is a molecular machine called
the proteasome, which both unfolds and chops superfluous proteins into smaller pieces
that will be used as raw building materials for new proteins. Given its critical role,
the proteasome is involved in multiple human diseases, and it serves as a perfect target
for a plethora of different drugs, most prominently, those commonly used in chemotherapy
of cancer. With the goal of understanding how these drugs work and paving the way for
designing even better drugs with less side effects, a
recent
collaborative study with
W. Baumeister (MPI Munich)
combined molecular dynamics flexible fitting (MDFF)
with de novo structure prediction algorithms to derive
four structural models from cryo-electron microscopy densities
of the proteasome in different conformational states. These models provide the first
atomic insights as to how ATP hydrolysis in the engine of the proteasome (cyan) unwinds
proteins and steers them towards the degradation chamber (red). More information is available on our
proteasome website. Easy access to our modeling techniques is
provided through QwikMD.