VMD-L Mailing List
From: Efthymiou, Christos (christos.dereschuk.20_at_ucl.ac.uk)
Date: Mon May 30 2022 - 14:08:16 CDT
- Next message: Vermaas, Josh: "Re: Clarification on RMSF Calculation"
- Previous message: Chris Taylor: "Re: Movie Maker plugin parallel MPI rendering"
- Next in thread: Vermaas, Josh: "Re: Clarification on RMSF Calculation"
- Reply: Vermaas, Josh: "Re: Clarification on RMSF Calculation"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]
Hello,
I have run a simulation on a protein-protein complex and I am using the QwikMD extension to run some analysis on the simulation.
When I calculate the RMSF vs Residue Number using the default selection of protein, I get RMSF values up to residue 352. I have two proteins in the simulation, one of which is numbered 1-294 and another 1109-1166. Based on this, I would assume residues 295-352 correspond to 1109-1166.
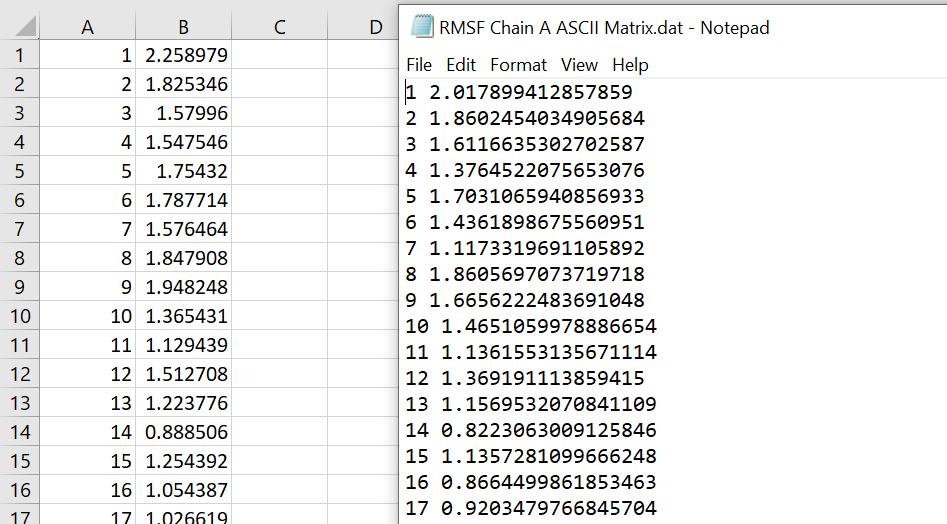
However, if I then calculate the RMSF with the selection of chain A (1-294) and chain C (1109-1166) separately, the numbers do not match the RMSF calculation for the protein selection. For example, on the left I have the RMSF calculation when protein was used as the selection and on the right is when chain A was used as the selection.
[cid:107a2645-af1c-424f-b094-0070c9b436ac]
I have also compared the RMSF calculation with selection of chain C compared to residues 295-352 from the protein selection, and those numbers also do not match.
Therefore, which selection is correct? And why do the numbers not match?
- Next message: Vermaas, Josh: "Re: Clarification on RMSF Calculation"
- Previous message: Chris Taylor: "Re: Movie Maker plugin parallel MPI rendering"
- Next in thread: Vermaas, Josh: "Re: Clarification on RMSF Calculation"
- Reply: Vermaas, Josh: "Re: Clarification on RMSF Calculation"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]



