Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
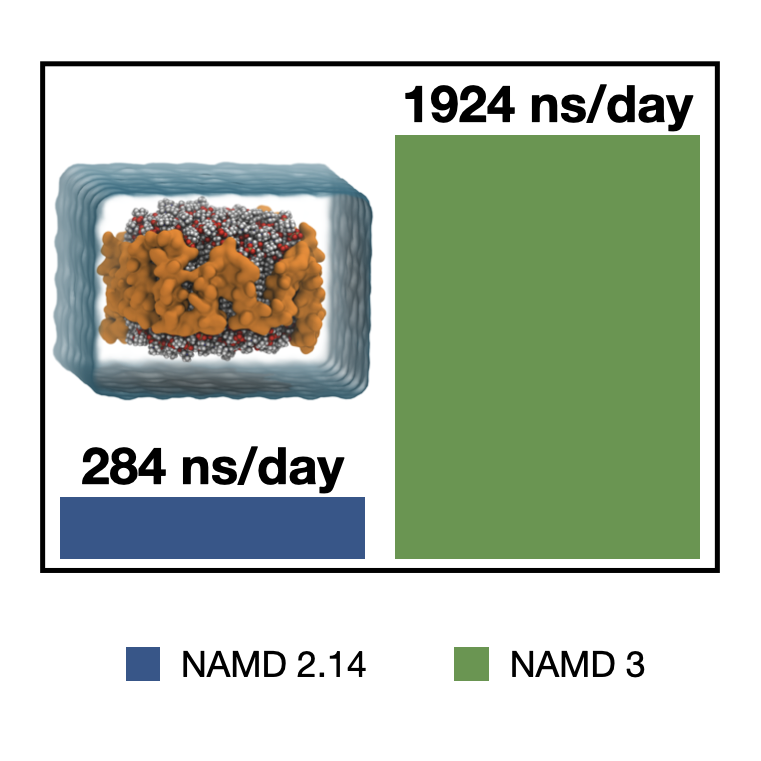
A major goal in molecular simulation, pursued by all programs for decades, is to reach faster simulations and longer timescales to describe, e.g., biological events more meaningfully. The Center has made a major stride in this direction in the recent critical update of NAMD, Version 3.0, which features a fully GPU-resident dynamics scheme for modern GPU architectures and eliminates many of the traditional NAMD overheads, such as its existing communication infrastructures, PCI latency, and the involvement of the CPU during integration, turning NAMD 3 into a completely GPU-bound application. These improvements allow NAMD 3 to perform up to 1.9x faster than v2.13 on existing Volta GPU architectures. It will be up to 1.4x faster on the upcoming NVIDIA A100. Finally, having no involvement of the CPU at all allows NAMD 3 scale perfectly for multi-copy simulation on multi-GPU systems. NAMD 3 performance will grow further as it matures, and as we begin to work on strong scaling with NVLink- and NVSwitch-connected GPUs for single-replica simulations in near future. See more information at most recent NVIDIA developer blog post and in the 2020 NAMD reference paper .