Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
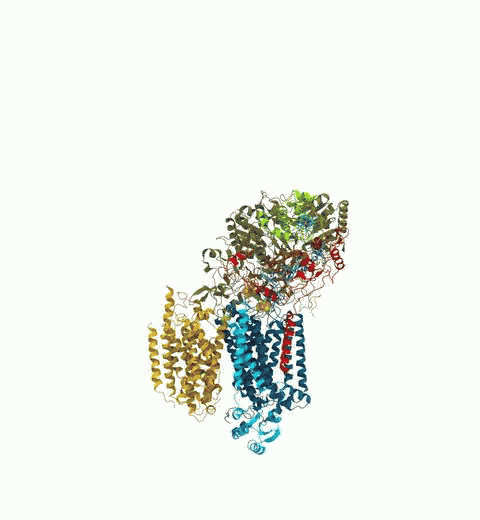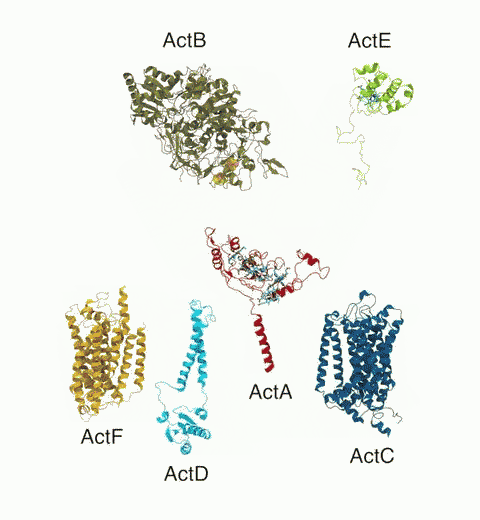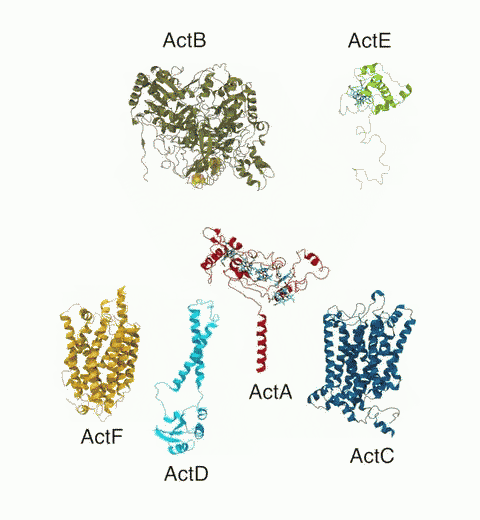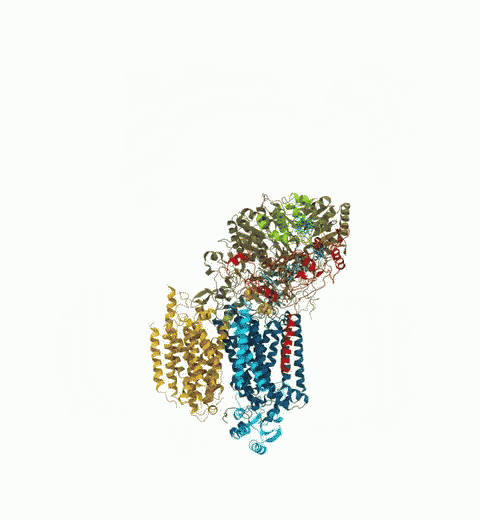
image size:
1.9MB
made with VMD
Bacteria are remarkable at tailoring their bioenergetic machineries to adapt to and thrive in diverse and ever-changing environments. A critical metabolic task is the efficient extraction of energy from food. Similar to us, many bacteria pass electrons to oxygen or other acceptors with the help of membrane-bound enzymes, and in doing so, they move protons across their membrane, thus generating a transmembrane voltage (much like a battery) that can be used for ATP synthesis. Typically, one enzyme passes an electron to its downstream enzyme via random collisions. However, sometimes these enzymes can form a supercomplex that positions the enzymes very close to each other and with the right pose for faster electron transfer and therefore more efficient energy conversion. In a recent paper reporting a three-way collaboration between biochemists, experimental structural biologists, and the Center researchers, the structure of one such supercomplex (termed Alternative complex III) was determined at an atomic resolution through a combination of cryoEM and advanced molecular modeling and simulation tools performed with NAMD and VMD. Read more about this study in Nature.



