Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
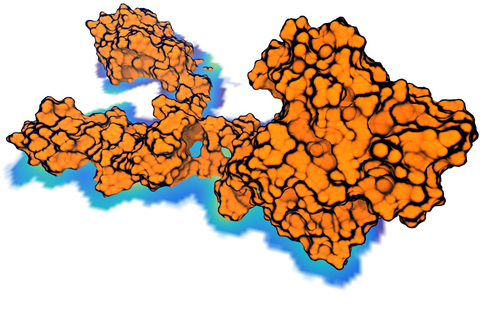
image size:
3.3MB
made with VMD
Living cells are brimming with the activity of macromolecular complexes carrying out
their assigned tasks. Structures of these complexes can be resolved
with cryo-electron microscopy (cryo-EM), wherein the complexes are first freeze-shocked into
states characterizing their action and subsequently imaged by detection cameras. Recent
advances in
direct detection camera technology enable today's cryo-EM laboratories to image the
macromolecular complexes at high-resolution, giving us a better view of the cell
than ever before. Computational techniques like molecular dynamics flexible fitting
(MDFF) are a key tool for producing atomic
models of the imaged molecules, providing greater insight into their structure and function.
The increased resolution of EM maps, which contain sharp valleys capable of trapping structures,
presents a challenge to
MDFF which was originally developed for maps in a
lower resolution range.
However, a recent study
unveils two new techniques called cascade (cMDFF) and
resolution exchange (ReMDFF) molecular dynamics flexible fitting to overcome the hurdles
posed by high-resolution maps. The refinement is achieved by interpreting a
range of cryo-EM images, starting with an image of fuzzy resolution and progressively
improving the image's contrast until near-atomic resolution is reached. These techniques
were employed to solve the structure of the
proteasome, the recycling machine of the human cell.
New analysis schemes that look at the flexibility of the obtained structure provide a
measure of model uncertainty within the near-atomic EM images, improving their contrast.
All the tools are
available on cloud computing platforms allowing community-wide usage at low
monetary cost; the complex computations can now be performed at the cost
of a cup of coffee.



