Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
NAMD
includes numerous enhancements to ..." />
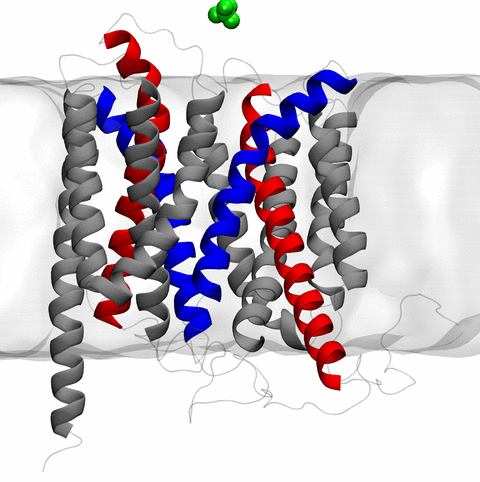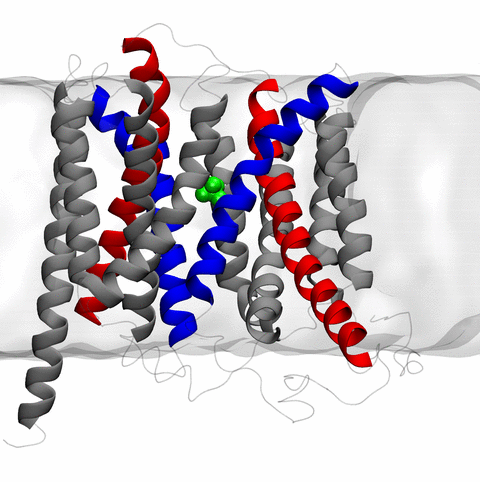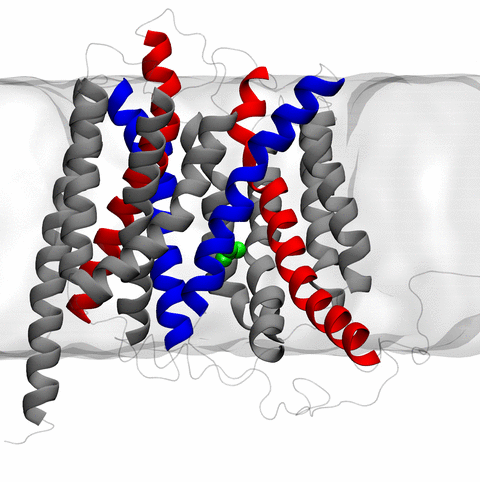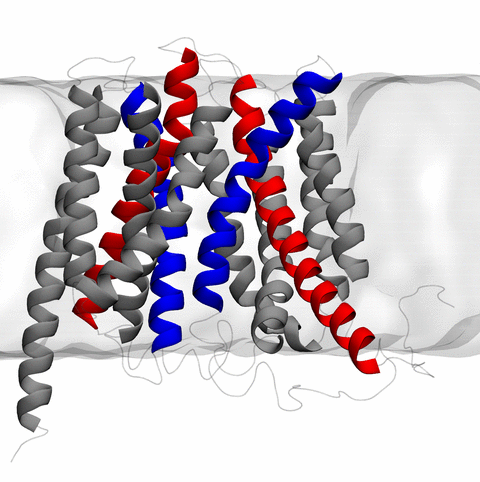

image size:
11.9MB
made with VMD
The 2.10 release of
the molecular dynamics program NAMD
includes numerous enhancements to support simulations
of massive supramolecular assemblies such as the HIV capsid (see highlight
Elusive HIV-1 Capsid).
A single such simulation of a hundred million atoms or more can
utilize the tens of thousands of processors of petascale supercomputers
thanks to recent advances in computational methodology
reported here.
Equally if not more significant, however, are advances in NAMD's
implementation of multiple copy algorithms for enhanced sampling of
smaller molecular systems as
reported here.
These algorithms have already allowed
researchers studying the molecular machinery of living cells
to reveal for the first time with NAMD
mechanisms operating on timescales of milliseconds or longer,
including
the rotary action of the ubiquitous energy conversion complex ATP synthase
and the inward to outward opening transition of
the membrane transporter protein GlpT, shown in the accompanying animation.
NAMD 2.10 also introduces multilevel summation,
reported here,
a major algorithmic advance
enabling efficient long-range electrostatics for non-periodic and semi-periodic simulations.
More on new features in the 2.10 release of NAMD can be found
here.



