Highlights of our Work
2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
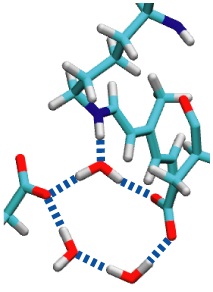
image size:
79.3KB
made with VMD
Sun light offers abundant energy fueling life on earth. Primary users of this energy are photosynthetic life forms. Remarkable in this regard are certain halophilic bacteria that developed a protein, bacteriorhodopsin (bR), which acting as a light driven proton pump turns sun light into a voltage gradient across the cell wall. Each photon absorbed primes bR to transfer of a proton. The bR pump had been discovered 30 years ago, yet the mechanism of the pump is still basically unknown. A key step towards establishing the mechanism would be identification of the initial energy form stored after light absorption. While some researchers suggest energy is stored highly localized in bR, others recognized that given the soft nature of proteins storage should be spread over many degrees of freedom. Widely accepted candidates are torsions of retinal, the linear molecule in bR that actually absorbs the light. A collaboration between computational and experimental groups carrying out unprecedented calculations and spectroscopic observations of water inside bR has now reported that a key fraction of light energy is stored in a geometrically distorted so-called hydrogen bond network involving retinal, three water molecules, and three amino acid side groups of bR. The results demonstrate dramatically that, in oder to reveal mechanisms underlying protein function, structural details at the sub-Angstrom level need to be resolved; only computational modeling guided by observation is presently capable of such resolution.



