Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
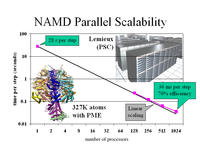
The parallel molecular dynamics program NAMD, and its sister visualization program VMD, have helped researchers at Illinois discern how muscles stretch, nerves sense pressure, and kidneys filter water. The latter project, for example, used simulations of 106,000 atoms to discover how aquaporins, which are ubiquitous in mammals, plants, and bacteria, allow water to pass while preventing the conduction of protons or ions (article in Science, more). Our years of work developing this software to apply the nation's fastest sup ercomputers to understand the tiny components of living cells were recognized at the SC2002 High Performance Networking and Computing conference with a Gordo n Bell Award for unprecedented parallel performance on a challenging computational problem (pdf of paper-497k). NAMD and VMD are distributed , free of charge, to thousands of scientists in industry and academia around the world, quickening the pace of drug discovery and other vital research to unravel biological processes.