Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
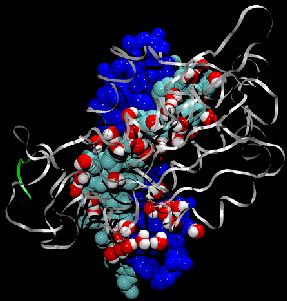
image size:
225.3KB
made with VMD
Deciphering the processes by which proteins recognize and bind to DNA is critical in our quest to understand cellular functions. To reach this goal, a collaboration with the group of Stephen Sligar, UIUC, explored the factors involved in protein-DNA recognition using hydrostatic pressure to perturb the binding of the BamHI endonuclease to cognate DNA. Our joint resulting publication outlines a new technique of high-pressure gel mobility shift analysis to test the effects of elevated hydrostatic pressure on the binding of BamHI (so-called restriction enzyme) to a specific DNA sequence. Upon application of a hydrostatic pressure of 500 bar, recognition between BamHI and the DNA sequence was weakened nearly 10-fold, suggesting an important role of water. An advanced 65,000 atom nanosecond molecular dynamics simulations with NAMD, at both ambient and elevated pressures, complemented the experiments and revealed how water-mediated interactions between BamHI and DNA control sequence recognition.



