VMD 1.8.5
The Theoretical and Computational Biophysics Group is pleased to announce
VMD version 1.8.5. Though only a minor version number different from the
previous release, many new structure building and analysis tools have been
added in this release make it easier for researchers to use
VMD in the process of setting up, running, and analyzing computer
simulations of biomolecules. Even more exciting, VMD
through an included plugin can now analyze multiple sequences,
closely integrating structure and sequence information.
Major features included in VMD 1.8.5:
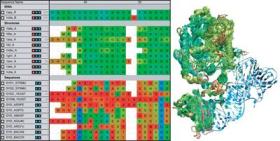
MultiSeq 2.0 Plugin
The MacOS X versions of VMD now support stereoscopic display on appropriately equipped machines (both Intel and PowerPC).
New and updated BioCoRE
collaboration plugins allow VMD users to
interactively chat,
submit jobs to supercomputers and clusters,
and exchange VMD views,
all without the need to to install any additional software.
An updated version of the APBS plugin now supports job remote execution through BioCoRE.
An updated version of the NAMDEnergy plugin now supports remote job execution
VMD includes new text commands and graphical interfaces for calculating
radial pair distribution functions,
IR spectral densities,
and
finding salt bridges.
New and updated structure building plugins provide text commands and
graphical interfaces for
structure building and editing,
and
converting between all-atom and coarse grain simulation models,
building PSF structure files,
and
calculating force field parameters.
The latest version includes support for several
additional file formats
through newly added molfile plugins.
A new
"Virtual DNA" visualization plugin
can display a graphical representation of DNA according to its helical
parameters (no atomic structure).
Short list of other popular features and improvements in VMD 1.8.5:
- NewCartoon representations now draw nucleic acid structures with ribbons for the backbone and cylinders for each nucleotide
- Pick points can now be created for user-defined graphics, to allow the creation of completely customized graphics and behaviors using the scripting interfaces.
- The built-in color table and menus now contain 32 user-modifiable base colors rather than the old limit of 16.
- Added new "RGryB" and "BGryR" color scales for use when coloring by potential on white backgrounds
- New representation cloning visualization plugin makes it easy to copy a set of representations from one molecule to another
- New Palette Tool visualization plugin displays active colors and materials, for test rendering and printing
- New NavFly mouse-based flying camera navigation plugin
- The Mutator plugin now includes support for free energy perturbation
- Improved the clipping plane tool user interface
- Updated Color Scale Bar plugin now has better range controls, improved behavior with white backgrounds, and generally more robust.
- Improved Ramachandran plotting plugin includes a new 3-D time averaged histogram display feature.
- New 'measure dipole' command for calculating dipole moments
- New 'measure gofr' command for calculating radial pair distribution functions
- New 'mol reanalyze' command allows one to force VMD to completely re-analyze the molecular structure due to changes in atom names, residue names, bonding/topology, etc. This is intended to be used by structure building tools and other special applications where one wishes to change the structure on-the-fly.
- Dynamic reclalculation of bonds can now be set for several atom-oriented representations, for use with trajectory playback
- Atom labels can now be displayed with user defined format strings, and with a defined offset from the atom coordinate.
- New VMDMACENABLEEEXTENSIONS environment variable enables performance-oriented OpenGL extensions which trigger bugs on some Mac systems.
- New VMDSHEARSTEREO environment variable enables an alternative perspective projection mode for improved stereoscopic display