Mutator Plugin, Version 1.3
This plugin provides a very simple method for mutating a target
residue selected by its segment name and id, and the three character
residue code for the mutant amino acid. It can also build, for the
specific purpose of free energy calculations, a hybrid structure where
both the initial and mutated side chains are present.
Normal operation
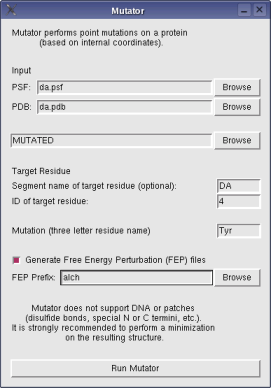
The plugin takes in psf and pdb files containing your protein (if you are not familiar with psf files, check out the autopsf plugin or the namd tutorial), the segment name and ID of the specific residue to be mutated (if you want to mutate residue 4 of segment DA in your protein, then Segment would be "DA" and ID would be "4"), and the final mutation (lets say you want to replace the amino acid at position 4 by tyrosine, then the three letter residue name for the mutation would be TYR). The mutated protein will be saved by default in files called "MUTATED.pdb" and "MUTATED.psf" on your working directory. If a segment is not specified, then the mutation will be performed on all protein segments of the system (note that mutator is able to handle systems that already include membrane, ions, and water).
Caveats
Mutator does not support DNA. Patches are supported provided they are listed in the REMARKS section of the input psf file. Since coordinates for the mutated amino-acid are assigned based on internal coordinates provided in standard CHARMM topology files, it is strongly recommended to perform an energy minimization on the resulting structure.
Making a hybrid with the FEP option
The FEP option is used in the specific case where you wish to study the thermodynamic impact of the mutation through free energy calculations with NAMD. This can be done using the alchemical Free Energy Perturbation (FEP) code. For alchemical FEP, you need a hybrid structure where both the original and the mutated amino acid side chains are present. When run with the FEP option, Mutator produces the files required for such a calculation. These include a fepFile and a PSF file modified to exclude all interactions between the original amino acid side chain and the mutated one.
Caveats for the FEP option
- Because of atom name changes, patches that modify the side chains cannot be used with the provided hybrid topologies. Standard patches for the termini should work fine, although it is always advisable to carefully inspect the resulting structure.
- In the particular case of glycine, the alpha carbon atom has to be modified in the transformation. For that reason, most patches will probably cause problems. Also, mutating a glycine will cause some angle and dihedral parameters to be duplicated, possibly modifying backbone conformational preferences. In short, do not mutate a residue from or to glycine unless you know what you are doing.
- Since proline has a special structure (and is actually not an
amino acid), hybrids involving proline are not supported.
- If the PSF produced by Mutator/FEP is further processed (by psfgen, solvate, ionize, etc.), the non bonded exclusion lists are likely to be lost. The resulting PSF should then be alchemified again to add these lists back.



