VMD-L Mailing List
From: parinaz bashirbanaem (parinaz.bashirbanaem_at_gmail.com)
Date: Fri Feb 19 2021 - 11:19:36 CST
- Next message: Chang, Christopher: "Re: Plotting orbitals from Molden file"
- Previous message: parinaz bashirbanaem: "question about RDF"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]
Hi all,
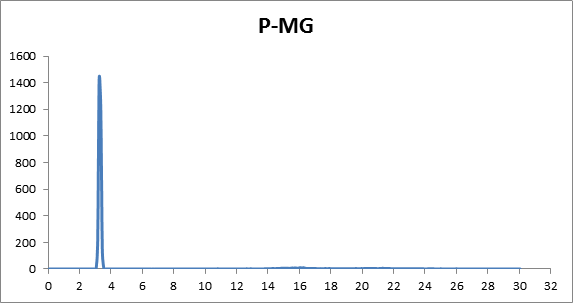
I am running MD simulation (NAMD package) for the DNA-RNA complex. I want
to calculate RDF by VMD, I have opened my trajectory by loading *.pdb
file and *.dcd file. For the calculation of the RDF, in the *selection-1
*section I have selected “nucleic and resid 51 and index 1598” and for
selection-2 section I have selected “resname MG”. However, I am not sure
that my process is right or not because the result is a bit anomalous from
the reference one.
Thank you very much for any suggestion.
Best regards,
Parinaz.
[image: image.png]
- Next message: Chang, Christopher: "Re: Plotting orbitals from Molden file"
- Previous message: parinaz bashirbanaem: "question about RDF"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]



