Simulation of a Photosynthetic Membrane
Chromatophores
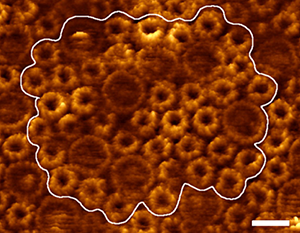
Fig. 1. An AFM image from the photosynthetic bacterium Rsp. photometricum was used to construct the photosynthetic membrane patch model.
Purple photosynthetic bacteria were likely the earliest photosynthetic organisms; in the present day, they can often be found at the bottom of lakes and ponds, where they make use of the green and far-red light not consumed by the plants and algae above. When these bacteria switch into photosynthetic mode, they begin producing light-harvesting proteins and reaction centers, and their inner cell membrane begins to change, budding out into specialized pseudo-organelles which will house the photosynthetic machinery. These structures, often referred to as "chromatophores" come in various shapes, such as tubules, long lamellar folds, or small vesicles.
AFM imaging of chromatophore membranes has provided some insight into the arrangement of the photosynthetic proteins within the chromatophore. Easiest to identify in these AFM images are the abundant light harvesting proteins, called light-harvesting complex 1 and 2 (LH1 and LH2). In species with lamellar (long flat folds) chromatophores (such as Ph. molischianum and Rsp. photometricum), AFM images have revealed domains containing a mix of LH1-RC monomers and LH2s, as well as LH2-only regions.

Fig. 2. Model of a photosynthetic membrane patch from a lamellar (flat) chromatophore.
Constructing a Model
A recent paper described how models were constructed representing a large portion of a membrane patch from Rsp. photometricum as seen in AFM images by Scheuring et al. (see here). Data from AFM regarding the locations and rotational orientations of the proteins in the membrane patch, were used as a starting point for building the models (a video showing the construction of the system can be seen here). Rigid structures of LH2s and LH1-RC monomers were fitted onto the AFM image. The initial placements of the proteins sometimes resulted in structural overlap between adjacent protein complexes, because the rigid placement could not account for distortion of the flexible LH1 rings, or the tilt of the LH1 and LH2 complexes in the membrane. These clashes were resolved by manually moving the complexes away from one another into a final position that appeared reasonable from the perspective of the overall packing density seen in the AFM image. The simulation system, including solvent, contained around 20 million atoms. The chromatophore membrane was equilibrated using the molecular dynamics software NAMD on petascale supercomputing resources including Titan (ORNL, via DOE INCITE), Blue Waters (NCSA), and Tsubame (Tokyo). The final model can be seen in Fig. 2. An artistic representation of the model overlaid onto the AFM image can be found here.
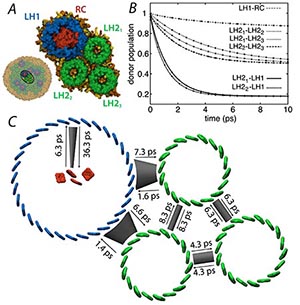
Fig. 3. (A) Subsystem comprising one LH1-RC and three LH2s to illustrate results of transfer rate calculations using the HEOM. (B) Relaxation of donor excited state population as calculated by the HEOM (lines) and using fitted transfer rates of a simple kinetic model (circles). (C) Excitation transfer network with excitation transfer times obtained from fitted transfer rates.
This chromatophore membrane simulation was used to study physical properties of the system, as well as to examine energy migration among the pigment molecules in the light harvesting complexes, eventually reaching the reaction center, where that energy is used to create a transmembrane charge separation, the first step in converting light energy eventually into chemical energy in the form of ATP for the cell. Quantum-dynamics calculations were then performed using the software PHI (see here) to determine a set of transfer rates that quantitatively characterize the intercomplex excitation transfer network, which was then used to calculate two key system characteristics - the excited state lifetime and the light harvesting efficiency of the system.
Excitation Transfer
Efficient excitation transfer between pigment-protein complexes is vital to efficient light harvesting. It has been shown that the excitation transfer strongly depends on intercomplex separation, and that intracomplex quantum coherence plays a vital role in excitation transport. Exciton transfer rates were determined using the hierarchy equations of motion (HEOM) that describe the dynamics of a quantum system in contact with a thermal environment; the actual separation between light harvesting proteins LH1 and LH2 was taken into account in the calculations.
A subsystem composed of one LH1-RC complex and three LH2 complexes, shown in Fig. 2 illustrates the determination of transfer rates from density matrix trajectories. Employing the transfer rates calculated via HEOM yields an exciton lifetime of 94.9 ps and a light harvesting efficiency of 90.5%.
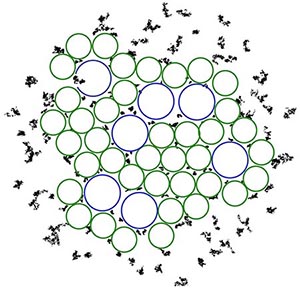
Fig. 4. Visualization of quinone (black dots) mobility in different local environments around the LH1 and LH2 complexes (blue and green circles).
Quinone Diffusion and Protein Packing
There are many photosynthetic processes that occur simultaneous to excitation transfer, such as the diffusion of quinones from LH1-RC to cytochrome bc1. While the dense packing of complexes in the chromatophore membrane favors rapid excitation transfer, it hinders quinone diffusion. The molecular dynamics simulations of the chromatophore membrane tracked the mobility of 150 quinone molecules placed in various parts of the membrane. The quinone molecules in the lipid-rich region outside of the protein complexes, or in the large, lipid-filled gaps between protein complexes, showed much more mobility than quinones placed in smaller gaps between proteins, as seen in Fig. 4.
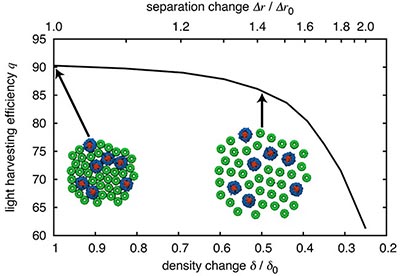
Fig. 5. Dependence of light harvesting efficiency on packing density. Reducing the packing density of light harvesting proteins, e.g., by a factor of 2, results in only a 4% decrease in light harvesting efficiency.
A question that naturally arises is whether the observed dense packing is a strict requirement for efficient light harvesting. The low computational cost of generalized Forster theory permits one to examine the effect of packing density on light harvesting efficiency. Fig. 5 shows this dependence. Reducing the light harvesting protein packing density by 50% results in only a 4% decrease in light harvesting efficiency.
Conclusions
The value of the procedures used in this investigation are that they not rely on prior assumptions of membrane organization or packing and yields the excitation transport network of a large system from low-resolution data. It is thus an attractive procedure to use in studying other, possibly more complex systems for which only low-resolution data on membrane organization is available. Using molecular-dynamics simulations as input to study quantum processes allows one to account for the hierarchy of physical interactions prevalent in biological systems.
Related publications
- Light harvesting by lamellar chromatophores in Rhodospirillum photometricum. Danielle Chandler, Johan Strumpfer, Melih Sener, Simon Scheuring, and Klaus Schulten. Biophysical Journal, 2014. In press.
- Integration of energy and electron transfer processes in the photosynthetic membrane of Rhodobacter sphaeroides. Michael L. Cartron, John D. Olsen, Melih Sener, Philip J. Jackson, Amanda A. Brindley, Pu Qian, Mark J. Dickman, Graham J. Leggett, Klaus Schulten, and C. Neil Hunter. Biochimica et Biophysica Acta - Bioenergetics, 2014. In press.
- Excited state dynamics in photosynthetic reaction center and light harvesting complex 1. Johan Strumpfer and Klaus Schulten. Journal of Chemical Physics, 137:065101, 2012. (8 pages).
- Open quantum dynamics calculations with the hierarchy equations of motion on parallel computers. Johan Strumpfer and Klaus Schulten. Journal of Chemical Theory and Computation, 8:2808-2816, 2012.
- Juxtaposing density matrix and classical path-based wave packet dynamics. Mortaza Aghtar, Jorg Liebers, Joha Strumpfer, Klaus Schulten, and Ulrich Kleinekathofer. Journal of Chemical Physics, 136:214101, 2012
- How quantum coherence assists photosynthetic light harvesting. Johan Strumpfer, Melih Sener, and Klaus Schulten. Journal of Physical Chemistry Letters, 3:536-542, 2012.
- The effect of correlated bath fluctuations on exciton transfer. Johan Strumpfer and Klaus Schulten. Journal of Chemical Physics, 134:095102, 2011. (9 pages).
- Forster energy transfer theory as reflected in the structures of photosynthetic light harvesting systems Melih ener, Johan Strumpfer, Jen Hsin, Danielle Chandler, Simon Scheuring, C. Neil Hunter, and Klaus Schulten. ChemPhysChem, 12:518-531, 2011.
- The light-harvesting apparatus in purple photosynthetic bacteria, introduction to a quantum biological device. Johan Strumpfer, Jen Hsin, Melih Sener, Danielle Chandler, and Klaus Schulten. In Benoit Roux, editor, Molecular Machines, chapter 2, pp. 19-48. World Scientific Press, 2011.
- Photosynthetic vesicle architecture and constraints on efficient energy harvesting. Melih Sener, Johan Strumpfer, John A. Timney, Arvi Freiberg, C. Neil Hunter, and Klaus Schulten. Biophysical Journal, 99:67-75, 2010.
- Self-assembly of photosynthetic membranes. Jen Hsin, Danielle E. Chandler, James Gumbart, Christopher B. Harrison, Melih Sener, Johan Strumpfer, and Klaus Schulten. ChemPhysChem, 11:1154-1159, 2010.
- Membrane curvature induced by aggregates of LH2s and monomeric LH1s. Danielle E. Chandler, James Gumbart, John D. Stack, Christophe Chipot, and Klaus Schulten. Biophysical Journal, 97:2978-2984, 2009.
- Protein-induced membrane curvature investigated through molecular dynamics flexible fitting. Jen Hsin, James Gumbart, Leonardo G. Trabuco, Elizabeth Villa, Pu Qian, C. Neil Hunter, and Klaus Schulten. Biophysical Journal, 97:321-329 (2009).
- Intrinsic Curvature Properties of Photosynthetic Proteins in Chromatophores. Danielle Chandler, Jen Hsin, Christophore B. Harrison, James Gumbart, and Klaus Schulten. Biophysical Journal, 95:2822-2836 (2008).
- From atomic-level structure to supramolecular organization in the photosynthetic unit of purple bacteria. Melih K. Sener and Klaus Schulten. In C. Neil Hunter, Fevzi Daldal, Marion C. Thurnauer, and J. Thomas Beatty, editors, The Purple Phototrophic Bacteria, volume 28 of Advances in Photosynthesis and Respiration, pp. 275-294. Springer, 2008.
Investigators
Collaborators
Simon Scheuring (INSERM, Aix-Marseille Universite)
Neil Hunter (University of Sheffield)
Related web pages on our site
Chromatophore and photosynthesis:
Structure of spherical chromatophore
Structure and function of the photosynthetic core complex
Atomic level structure and function of a photosynthetic vesicle
Organization of energy transfer networks.



