Research Topics - Nanoengineering
Molecular modeling provides nanoscale images at atomic and even electronic resolution, predicts the nanoscale interaction of yet unfamiliar combinations of biological and inorganic materials, and can evaluate strategies for redesigning biopolymers for nanotechnological uses. The methodology's value has been reviewed for three uses in bionanotechnology. The first involves the use of single-walled carbon nanotubes as biomedical sensors where a computationally efficient, yet accurate description of the influence of biomolecules on nanotube electronic properties and a description of nanotube - biomolecule interactions were developed; this development furnishes the ability to test nanotube electronic properties in realistic biological environments. The second case study involves the use of nanopores manufactured into electronic nanodevices based on silicon compounds for single molecule electrical recording, in particular, for DNA sequencing. Here, modeling combining classical molecular dynamics, material science, and device physics, describes the interaction of biopolymers, e.g., DNA, with silicon nitrate and silicon oxide pores, furnishes accurate dynamic images of pore translocation processes, and predicts signals. The third case involves the development of nanoscale lipid bilayers for the study of embedded membrane proteins and cholesterol. Molecular modeling tested scaffold proteins, redesigned lipoproteins found in mammalian plasma that hold the discoidal membranes in shape, and predicted the assembly as well as final structure of the nanodiscs. In entirely new technological areas like bionanotechnology qualitative concepts, pictures, and suggestions are sorely needed; the three exemplary applications document that molecular modeling can serve a critical role for the new bionanotechnology, even though it may still fall short on quantitative precision.

image size:
268.6KB
made with VMD
image size:
91.7KB
made using VMD
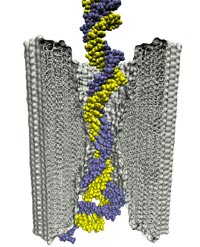
The most celebrated molecule of living cells, DNA, owes its fame to its role as a carrier of genetic information. But DNA is also impressive through other amazing properties, for example its mechanical flexibility. At first sight, it might seem a dull question to ask what is the smallest pore DNA can be squeezed through, as the obvious answer is that the diameter of that pore should be slightly larger than the diameter of a DNA helix. However, recent studies (paper1, paper2) in asking the stated question discovered that double stranded DNA can permeate, without loosing its structural integrity, pores smaller in diameter than a DNA double helix. The discovery was initiated through molecular dynamics simulations, carried out using NAMD and VMD. The simulations demonstrated that if an electrical field, driving negatively charged DNA through a nanopore, exceeds some critical value, the force exerted on DNA stretches DNA to twice its equilibrium length, reducing thereby its diameter and allowing it to squeeze through narrow pores. The simulations predicted precise values of pore radii and associated critical fields. The predictions were validated experimentally by counting the number of DNA copies that passed at different electric fields through synthetic nanopores. Further details about this study can be found here.
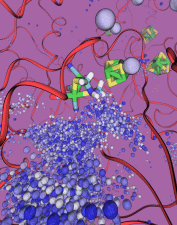
image size:
102.1KB
made with VMD
In an optimistic future, cars and appliances will be powered by renewable energy produced by burning hydrogen gas, with water being the only waste product. To supply this hydrogen gas, scientists are turning their attention to an enzyme called hydrogenase that is found in certain microorganisms, which produce hydrogen gas from sunlight and water. This enzyme, however, is sensitive to oxygen gas, which irreversibly deactivates its hydrogen-producing active site. Understanding how oxygen reaches the active site will provide insight into how hydrogenase's oxygen tolerance can be increased through protein engineering, and in turn make hydrogenase an economical source of hydrogen fuel. In a recent paper (also described in this webpage), the programs NAMD and VMD are used to analyze the gas diffusion process inside hydrogenase, and how it correlates with the protein's internal fluctuations, thereby creating a map of the oxygen pathways. The calculations revealed two distinct pathways for oxygen to reach the active site. Gases participate in physiological processes of many organisms and the new computational strategy developed promises to image gas diffusion pathways for many relevant proteins. In fact, the researchers are currently inspecting hundreds of proteins for their ability to internally transport gas molecules.
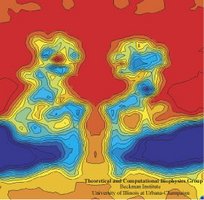
In a biological cell, membrane channels act like miniature valves regulating the flow of ions and other solutes between intracellular compartments and across the cell's boundary. Assembled in complex circuits, they generate, transmit, and amplify signals orchestrating cell function. To investigate how membrane channels work, life scientists, using an extremely fine pipette, isolate a tiny patch of a cell membrane and, in so-called patch clamp measurements, determine electric currents in response to applied electric potentials. Dramatic increase in computational power and its efficient utilization by NAMD allows one today to reproduce such studies computationally, calculating the permeability of a membrane channel to ions and water directly from its atomic structure. In what is one of the largest molecular dynamics simulation to date, described in a recent paper as well as on our web site (here), one copy of the membrane channel alpha-hemolysin, submerged in a lipid membrane and water, was subject to an external electric field that drove ions and water through the channel. The calculations produced also an image of the electrostatic potential across the channel (see figure).

image size:
117.6KB
made with VMD
Carbon nanotubes are becoming universal tools and building blocks
in nanomedicine, being proposed as nanodevices for drug delivery,
DNA transfection, and biosensing. They can also be employed as
nanopores that conduct protons, ions, and small molecules (see June 2003 highlight), or
as reaction vessels for new types of chemical reactions. Studying
carbon nanotubes can assist in designing improved nanodevices. One
approach to studying nanotubes is furnished by molecular dynamics
simulations. However, such simulations must account for one key
property of nanotubes, their large polarizability due to the
mobility of ![]() -electrons over the
tube walls. Until recently this polarizability could only be calculated
through expensive quantum chemical calculations that could not be linked
to simulations imaging molecular processes around carbon nanotubes.
Recent studies (
1 ,
2 )
reports now an empirical model that can be efficiently implemented into
molecular dynamics simulations to take into account the polarization
effect. The model reproduces results of more expensive quantum chemistry
calculations very well. A first application of the new model studied
the transport of water molecules through nanotubes. Water has a strong
dipole moment that polarizes the nanotube wall and, therefore, provides a
stringent test for the new methodology.
-electrons over the
tube walls. Until recently this polarizability could only be calculated
through expensive quantum chemical calculations that could not be linked
to simulations imaging molecular processes around carbon nanotubes.
Recent studies (
1 ,
2 )
reports now an empirical model that can be efficiently implemented into
molecular dynamics simulations to take into account the polarization
effect. The model reproduces results of more expensive quantum chemistry
calculations very well. A first application of the new model studied
the transport of water molecules through nanotubes. Water has a strong
dipole moment that polarizes the nanotube wall and, therefore, provides a
stringent test for the new methodology.
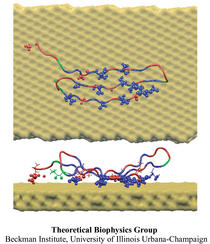
image size:
91.6KB
made with VMD
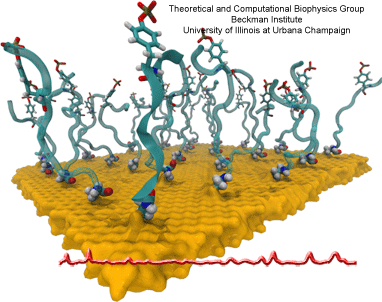
The biological control of inorganic crystal formation, morphology and assembly is of interest to biologists and biotechnologists studying hard tissue growth and regeneration, as well as to materials scientists using biomimetic approaches for control of inorganic material fabrication and assembly. A molecular-level understanding of the natural mechanisms involved in these processes can be derived from the use of metal surfaces to study surface recognition by proteins together with combinatorial genetics techniques for selection of suitable peptides.
In a recent study, the structure of a genetically engineered gold binding protein has been determined computationally, and the ability of the protein to recognize gold crystal surfaces has been explained. Molecular dynamics simulations were carried out with the program NAMD using the solvated protein at the gold surface to assess the dynamics of the binding process and the effects of surface topography on the specificity of protein binding.
Nanopores have emerged as promising next generation devices for single-molecule detection, analysis and DNA sequencing. One of the drawbacks of solid-state nanopores is that the membrane thickness is about 100 times the distance between two bases in a DNA molecule. Graphene is a material with extraordinary electrical and mechanical properties. It is the thinnest known material with thickness equal to one atomic layer of carbon (0.3 nm), which is comparable to the DNA base pair stacking distance of 0.35 nm, making the graphene nanopore a promising device for DNA sequencing.
Detection of chemically active molecules remains a challenge for biomedical applications. Ideally, current miniaturization technology should allow manufacturing very small devices, nanodevices, to detect enzyme activity. Indeed, a long-awaited breakthrough would be the advent of kinase profiling microarrays. Protein kinases are known to regulate the majority of biochemical pathways in the cell, controlling a variety of functions such as cell growth, differentiation and mobility. The main function of kinases is to activate other proteins by phosphorylation; that is, to transfer a phosphate group (PO4) from ATP to a sidechain, usually a serine, threonine, or tyrosine. Disruptions of kinase signaling pathways are frequent causes for diseases, such as cancer and diabetes. Therefore, a sensitive tool to accurately detect phosphorylation is of central importance for biomedical research.
Nanopores are incredible small pores with diameters of just a few nanometers, fabricated on silica (SiO2) or other synthetic materials. They can be used to study the translocation of charged species, such as ions or charged molecules. Since, most nanopore applications are based on measuring the changes in the ionic current, understanding the ion dynamics through nanopores is desirable. Indeed, ionic current experiments with nanopores still surprise researchers, many fascinating results have been reported, such as ionic current rectification, nanobubbles, nanoprecipitation and surface charge inversion. Here, we present a summary of our studies about ionic current rectification in silica nanopores.