Structure and Function of the Photosynthetic Core Complex
Light absorption and the subsequent transfer of excitation energy are the first two steps of photosynthesis. In photosynthetic bacteria, these two steps are carried out in the large membrane protein assembly called the photosynthetic core complex. There are two main constituent proteins in the core complex: light-harvesting complex I (LH1) and the reaction center (RC). The LH1 performs the task of absorbing light energy and transferring the energy to a RC, which then utilizes this energy to produce a charge difference across the cellular membrane, driving ATP synthase to convert ATP.
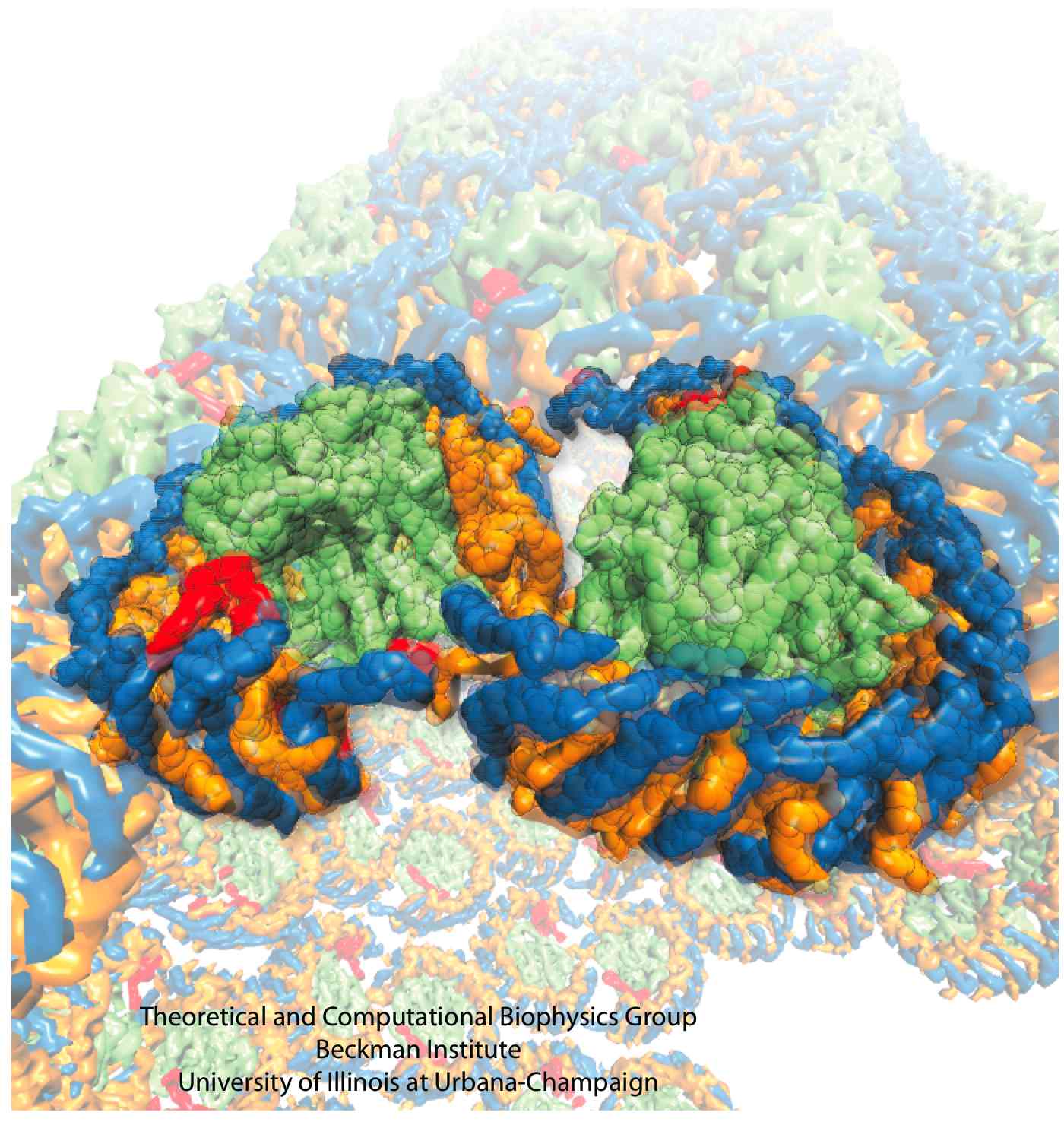
In different bacterial species, the photosynthetic core complex can take on very different organizations. Typically, the LH1 subunits surrounds directly the slightly elliptical RC. In some cases, the LH1 forms a complete ring, an example being Rhodospirillum rubrum (Figure 1A). The core complexes of certain species contain an additional, single transmembrane alpha-helical protein. In Rhodopseudomonas palustris, this extra protein is called the W protein, and the LH1 ring is seen to exhibit a gap where the W protein resides (Figure 1B). In the Rhodobacter species, there is a similar extra protein called PufX, which causes the core complex to dimerize (Figure 1C and D) in Rhodobacter sphaeroides.

Figure 1 - Schematics of different organizations of the bacterial photosynthetic core complexes. (A) A core complex in which the LH1 subunits form a complete ring, as seen in Rhodospirillum rubrum. (B) A core complex in which the LH1 subunits forms a ring with a gap, with an extra polypeptide near the gap, as seen in Rhodopseudomonas palustris. (C) and (D) Two proposed organizations for a dimeric core complex, the dimerization of which requires the extra polypeptide, PufX. (C) is drawn according to the highest structural data for a dimeric core complex (Qian et al., 2005), which places PufX near the LH1 gaps. In (D), PufX is assumed to dimerize and is situated at the center of the core complex. Dimeric core complexes are seen in certain Rhodobacter species, the best-known case being Rhodobacter sphaeroides.
A structural model of a dimeric core complex and its light-harvesting properties
The light-absorption function of the LH1 is carried out by pigments (bacteriochlorophylls, abbreviated as BChls) embedded in the protein. After absorbing light energy, the pigments pass the excitation energy around, and the light energy is eventually funneled to the RC. The rate of such energy transfer is dependent on the geometry of the pigment organization, which in turn is defined largely by protein geometry, since pigments are fixated at certain preferred binding sites. With a precise knowledge of where the pigments are, theoretical frameworks are available to compute the excitation migration between different LH1 pigments, as well as the transfer of excitation between LH1 and RC.
Since no high-resolution structure is available for a complete core complex, some modeling efforts are necessary. We have constructed a rudimentary core complex model for the species Rhodobacter sphaeroides, whose core complex contains PufX and is dimeric (Figure 1C and D) (see Chandler et al., 2008). With the publication of the first three-dimensional structural data of Rhodobacter sphaeroides core complex dimer at low resolution (Qian et al., 2008), the model was further improved (see Sener et al., 2009 and Hsin et al., 2009a). The atomic model of the core complex (arranged in the organization shown in Figure 1C) provides a starting point for understanding how excitations are passed among the pigments in LH1 and the RC; results of the calculation is summarized in Figure 2, with details available in Sener et al., 2009.

Figure 2 - Structural model of the Rhodobacter sphaeroides core complex and excitation states. (A) Setup of the all-atom simulation that combined modeling with the low-resolution structural data of the Rhodobacter sphaeroides core complex. (B) The positions of the BChl pigments before the simulation. Pigments are shown in purple, with the low-resolution structural data in transparent gray. (C) Organization of the pigment can be seen to adopt to the large bent of the low-resolution data. (D) Close-up view of the pigments. (E) An example of the excitation properties of the Rhodobacter sphaeroides core complex dimer calculated in
Sener et al., 2009. Shown here is the spread of the lowest exciton states over the BChl array of the dimeric complex. (A-D) made with VMD.
The Rhodobacter sphaeroides core complex is capable of bending the cellular membrane. This process is captured in a molecular dynamics simulation, shown in the movie below:
Testing the dimerization of PufX
It is still not completely established where the PufX protein is in the Rhodobacter sphaeroides core complex dimer. While the highest structural data for the complex (Qian et al., 2005) suggested an organization depicted in Figure 1C, there are also imaging data interpreted as Figure 1D (for a comprehensive review, see Holden-Dye et al., 2008).
The organization shown in Figure 1D would require the direct association of the two PufX proteins, yet, notably, only monomeric solution structures of PufX have been reported. Busselez et al., 2007 speculated that the GXXXG (Glycine-anything-anything-anything-Glycine) motif in the PufX sequence provides a site of PufX-PufX self-association similar to that observed in the well-known dimeric membrane protein, glycophorin A. This hypothesis was put to a series of computational tests, reported in Hsin et al., 2009b and summarized in Figure 3.

Figure 3 - Molecular dynamics simulation of a PufX dimer. (A) Set up of the simulation with two PufX proteins, one in blue and one in orange. Membrane and water are both included in the system. The modeled PufX dimer stayed closely associated throughout the 50-ns simulated. At the end of the simulation, a uniform crossing angle between the two PufX helices developed, as traced in (B). (C) The association of the PufX helices can be estimated quantitatively by measuring the buried molecular surface area between the two proteins. The two PufX helices during the simulation remained closely in contact. (D) Residue-residue contact map identifying key residues that held together the PufX dimer model in simulation.
Molecular dynamics simulations showed that the modeled PufX dimer is structurally stable over the simulated 50 ns. If PufX indeed is positioned at the center of the Rhodobacter sphaeroides core complex dimer, then the molecular interactions between PufX and the LH1 subunits would lead to strucrual consequences in the overall geometry of the core complex. One feature considered in Hsin et al., 2009b was the effect of the PufX crossing angle. As LH1 subunits attach to the two sides of the PufX dimer forming two incomplete rings, the crossing angle in the PufX helices would lead to the two LH1 rings being on a different plane, with the angle made by the two planes related to the crossing angle of PufX approximately (Figure 4). We note that the definitive placement of PufX would require a high resoultion structure of the core complex.

Figure 4 - Potential structural consequences on the overall geometry of the core complex due to the dimerization of PufX. (A) A tentative model of the core complex following the organization depicted in Figure 1D with a PufX dimer at the center of the complex. (B) Crossing angle between the PufX helices can be translated to the overall bending angle of the core complex geometrically.
A movie illustrating a hypothesized scenario of core complex self-assembly can be started here.
A model of the Core Complex-Only Photosynthetic Membrane
In the absence of the light-harvesting complex II, the core complex can aggregate into orderly cylindrical vesicles with a diameter of tens of nanometers. In such cylindrical photosynthetic membrane, since only core complexes are present and they are arranged helically, it is possible to place each core complex and model the full membrane. Such model is shown in Figure 5.

Figure 5 - The tubular membrane model and its periodic architecture, the model constructed in Hsin et al., 2010. (a) The all-atom model of the helically arranged core complex dimers forming a tubular photosynthetic membrane. Full complexes are shown on the left half of the tube, with only the BChl arrays shown on the right half to illustrate the long-range organization of multiple BChl arrays. (b) Network defined by connecting the symmetry axes of core complexes constituting the membrane. The projection of this network onto a plane results in the periodic tiling shown in (c).
The model of the tubular photosynthetic membrane shown in Figure 5a would be precise if there is a full structure of the core complex. However, as discussed above, there are several structural aspects of the photosynthetic complex that are still under investigation, one example being the location of the PufX protein. Another structural uncertainty is exactly how many BChls are there per core complex dimer. The BChls absorb the light energy, and an excited BChl passes its energy to nearby BChls. This process of energy transfer has been described by various physical theories, with one popular choice being the Förster theory, and another being the computationally more costly, yet remarkably accurate treatment of the hierachical equation of motion, discussed and compared here. Both theoretical methods require the position of all BChls as an input, and, therefore, uncertainties in the BChl location translates into imprecision in the description of how light energy goes from one BChl to another.
To systematically determine how much a given alteration in BChl arrangements would affect the overall energy transfer dynamics in the cylindrical photosynthetic membrane, we carried out calculations on a variety of BChl arrays reflecting our current knowledge of the core complex structure. Also, both the Förster theory and the hierachical equation of motion were used for comparison, much like what was done in Strumpfer and Schulten, 2009, for light-harvesting complex 2. Our results showed that our large-scale understanding of the core complex structure is sufficient in computing the energy transfer characteristics of the core complex dimer, with the uncertainties in the structural details not changing significantly the calculation results. Also, much like reported in Strumpfer and Schulten, 2009, the Förster theory was shown to provide sufficiently accurate results.
Measuring the dimerization affinity of PufX from different bacteria
As shown in Figure 1 above, core complexes exist as monomers and dimers depending on bacterial species. While PufX is found in Rhodobacter species, this does not mean that all Rhodobacter bacteria have dimeric core complex. In fact, imaging studies have shown that Rhodobacter veldkampii only has monomeric, ring-shaped core complex, even though PufX is also present in this bacterium. So what determines the oligomerization state of core complexes, and how does the potential dimerization of PufX effect the assembly of core complexes? To address this question, computation was combined with biochemistry techniques, and the dimerization affinity of PufX from four different Rhodobacter bacteria was measured.

Figure 6 - Computationally measure the dimerization affinity of PufX from four different Rhodobacter bacteria. The PufX proteins tested included Rhodobacter blasticus (which has dimeric core complexes), Rhodobacter spheroids (which also has dimeric core complexes), Rhodobacter capsulatus (which is suspected to have dimeric core complexes), and Rhodobacter veldkampii (which only has monomeric core complexes).
Figure 6 above portrays typical simulation setups to probe the reversible process of PufX-PufX association and disassociation. Among the four bacterial species considered here, Rhodobacter veldkampii is the only one that is confirmed to have only monomeric PufX. Through such simulations, one can calculate the potential of mean force along the dimerization pathway, parametrized by the distance between the two PufX helices, as shown in Figure 7 below.

Figure 7 - Potential of mean force plots for the free-energy calculations shown in Figure 6. (a) and (b) show different set of simulations using slightly different transmembrane PufX segment, with the sequence included in the simulation highlighted in the inset. As can be seen, Rhodobacter veldkampii PufX consistently shows the lowest preference for dimerization.
The free-energy calculations suggest that Rhodobacter veldkampii PufX has nearly no affinity for dimerization, where as the other three Rhodobacter PufX showed some capability of dimerization. This trend was also observed in biochemical experiment (Figure 8), where the dimerization affinity of the same four PufX proteins was measured using a method known as TOXCAT. These results showed that bacteria with a PufX prone to dimerization also have dimeric core complex, and bacteria with a PufX incapable of dimerization have monomeric core complex. The core complex organization model shown in Figure 1D is the most consistent with these data.

Figure 8 - (a) Measuring dimerization affinity of PufX experimentally using TOXCAT. (b) TOXCAT results suggest that Rhodobacter capsulatus, Rhodobacter blastica, and Rhodobacter spheroids all have some tendency for dimerization, but Rhodobacter veldkampii is nearly incapable of dimerization. These results agree with the computational measurement, shown in (c).
Related publications
Oligomerization state of photosynthetic core complexes is correlated with the dimerization affinity of a transmembrane helix, Jen Hsin, Loren LaPointe, Alla Kazy, Christophe Chipot, Alessandro Senes, and Klaus Schulten. Journal of the American Chemical Society, 133:14071-14081 (2011).
Energy transfer dynamics in the RC-LH1-PufX core complex-only tubular photosynthetic membrane, Jen Hsin, Johan Strumpfer, Melih Sener, Pu Qian, C. Neil Hunter and Klaus Schulten. New Journal of Physics, 12:085005 (2010).
Self-assembly of photosynthetic membranes,
Jen Hsin, Danielle E. Chandler, James Gumbart, Christopher B. Harrison, Melih Sener, Johan Strumpfer and Klaus Schulten.
ChemPhysChem, 11:1154-1159 (2010).
A glycophorin A-like framework for the dimerization of photosynthetic core complexes.
Jen Hsin, Chris Chipot, and Klaus Schulten.
Journal of the American Chemical Society, 131:17096-17098 (2009).
Protein-induced membrane curvature investigated through molecular dynamics flexible fitting.
Jen Hsin, James Gumbart, Leonardo G. Trabuco, Elizabeth Villa, Pu Qian, C. Neil Hunter, and Klaus Schulten.
Biophysical Journal, 97:321-329 (2009).
Structural model and excitonic properties of the dimeric RC-LH1-PufX complex from Rhodobacter sphaeroides.
Melih Sener, Jen Hsin, Leonardo G. Trabuco, Elizabeth Villa, Pu Qian, C. Neil Hunter, and Klaus Schulten.
Chemical Physics, 357:188-197 (2009).
Intrinsic Curvature Properties of Photosynthetic Proteins in Chromatophores.
Danielle Chandler, Jen Hsin, Christophore B. Harrison, James Gumbart, and Klaus Schulten.
Biophysical Journal, 95:2822-2836 (2008).
Related Research Highlights
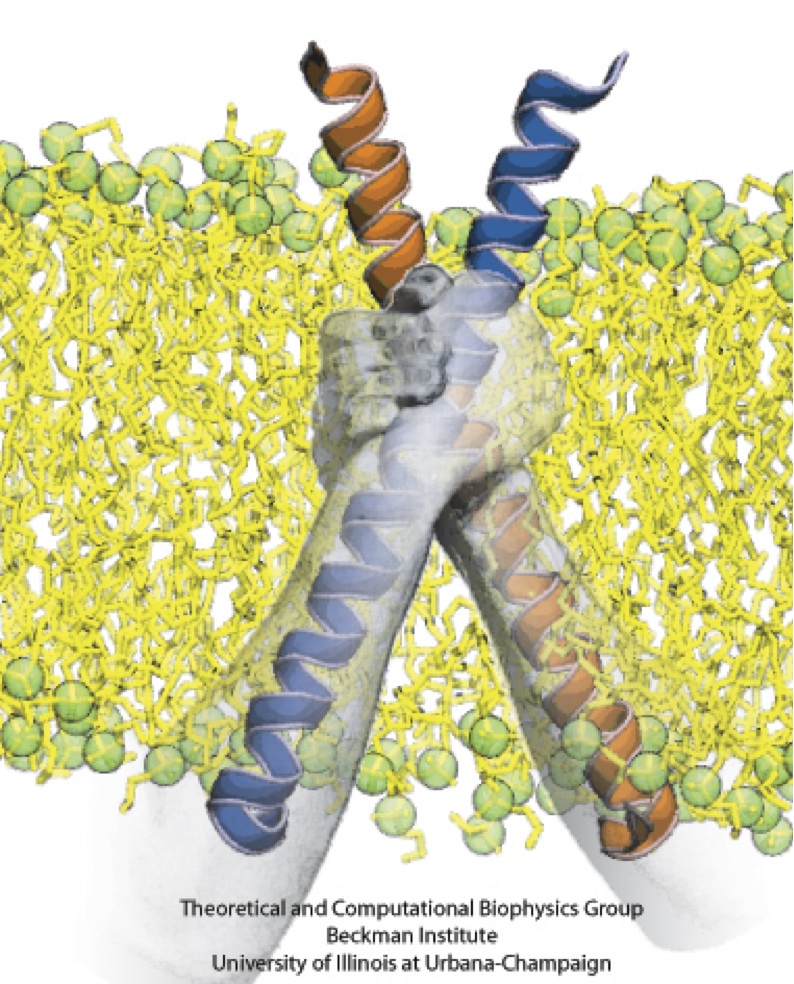
image size:
284.7KB
made with VMD
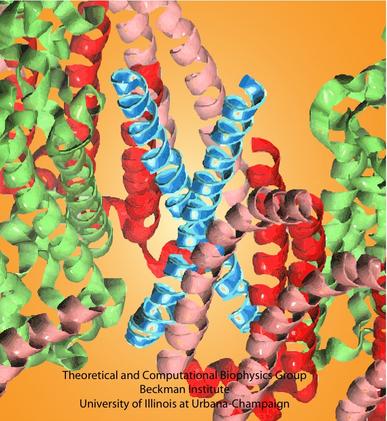
image size:
115.2KB
made with VMD
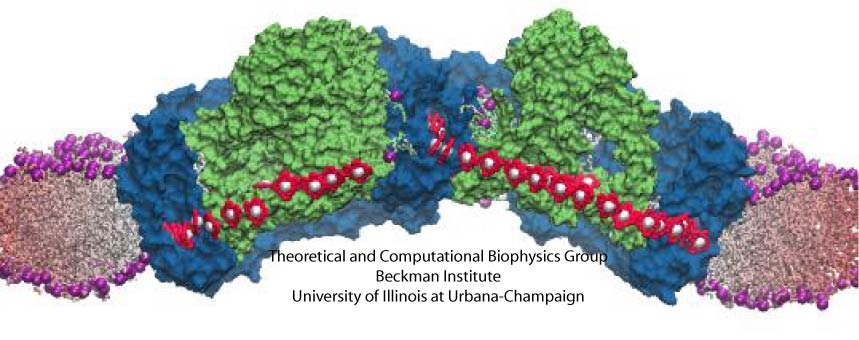
image size:
440.0KB
made with VMD
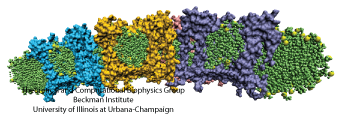
image size:
360.9KB
made with VMD
Investigators
Collaborators
Pu Qian (University of Sheffield)
Neil Hunter (University of Sheffield)
Related web pages on our site
Chromatophore and photosynthesis:
Intrinsic curvature properties of photosynthetic proteins in chromatophores
Atomic level structure and function of a photosynthetic vesicle
Organization of energy transfer networks.
Quantum Biology of the Photosynthetic Unit
Light-Harvesting by Carotenoids
Page created and maintained by Jen Hsin.