Chris Chipot

CNRS research director Adjunct professor of physics Contact information
Alternate address. Laboratoire international associé CNRS-UIUC. UMR 7565. Université de Lorraine. B.P. 70239. 54506 Vandœuvre-lès- Nancy, France Web: http://www.lia-uiuc.cnrs.fr/ |
Education
- MS at Université Henri Poincaré, France.- PhD at Université Henri Poincaré, France.
- Postdoctoral fellow at the Department of pharmaceutical chemistry, UCSF.
- Postdoctoral fellow at the NASA Ames Research Center.
Research areas
| Development | |||
| |||
| Applications |
Membrane proteins.
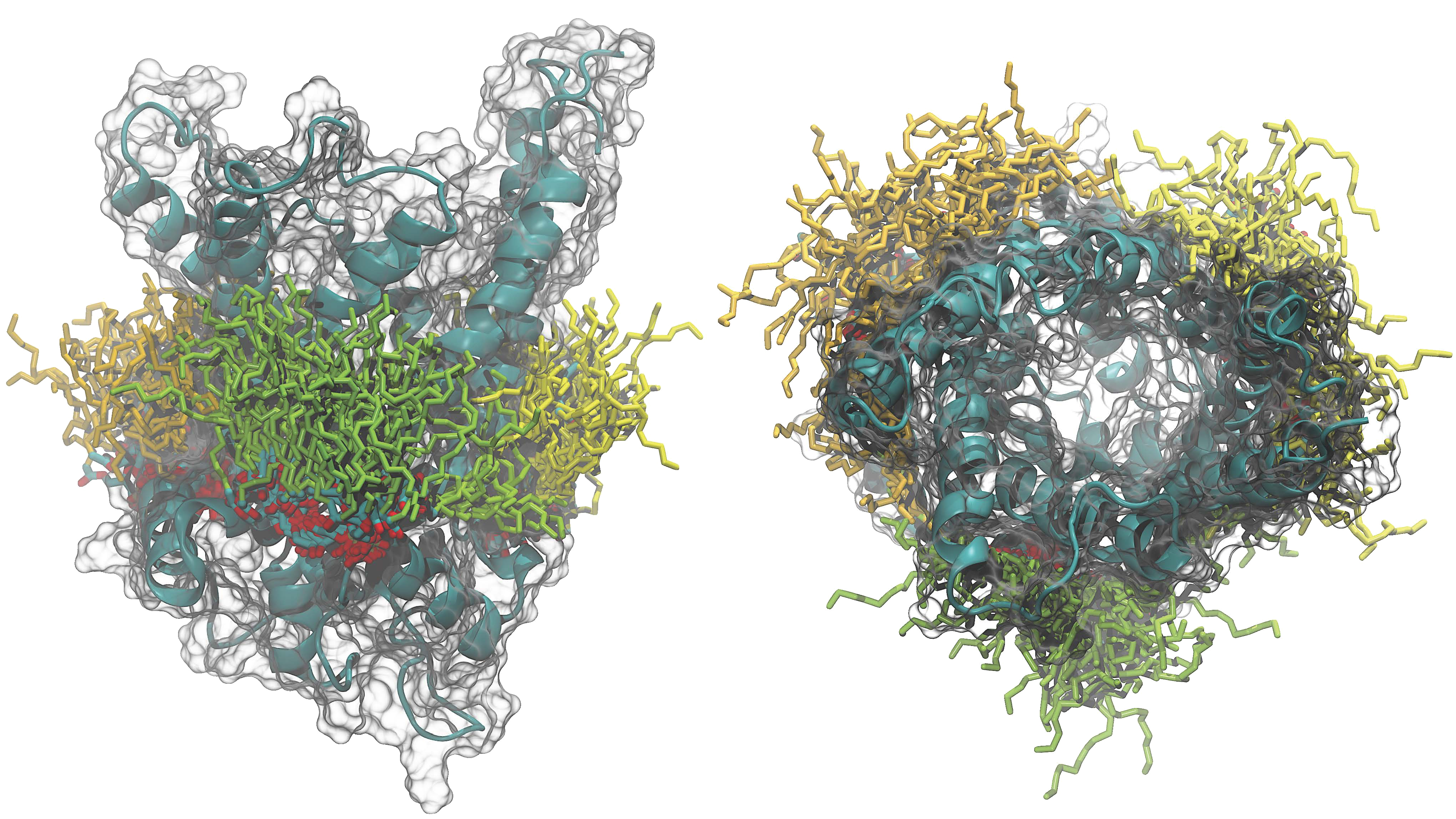 Membrane proteins are the gateways to the cell and to cellular
compartments. In combination with their sophisticated environment, they
perform a host of functions ranging from signal transduction, transport
of metabolites to energy conversion. In mammals, up to 30% of the
genome encodes membrane proteins, which today, represent the primary
target for drug discovery. Yet, notwithstanding their significant
population in the cell wall and their importance in cellular processes,
membrane proteins are markedly less well characterized than
hydrosoluble ones. This imbalance can be rationalized by the strong
dependence of the structure and stability of membrane proteins on their
native lipid environment. For many years, we have used
molecular-dynamics simulations in synergy with experiment to dissect
the interplay of membrane proteins with their surroundings, focusing on
members of the mitochondrial carrier family, and on the p7 protein of
Hepatitis C virus, crucial for assembly and release of infectious
virions. In addition to addressing the dynamics of membrane proteins in
near physiological conditions, we employ molecular dynamics simulations
to rationalize the information gleaned in structural biophysics
experiment in mimetic environments, notably in detergents.
Membrane proteins are the gateways to the cell and to cellular
compartments. In combination with their sophisticated environment, they
perform a host of functions ranging from signal transduction, transport
of metabolites to energy conversion. In mammals, up to 30% of the
genome encodes membrane proteins, which today, represent the primary
target for drug discovery. Yet, notwithstanding their significant
population in the cell wall and their importance in cellular processes,
membrane proteins are markedly less well characterized than
hydrosoluble ones. This imbalance can be rationalized by the strong
dependence of the structure and stability of membrane proteins on their
native lipid environment. For many years, we have used
molecular-dynamics simulations in synergy with experiment to dissect
the interplay of membrane proteins with their surroundings, focusing on
members of the mitochondrial carrier family, and on the p7 protein of
Hepatitis C virus, crucial for assembly and release of infectious
virions. In addition to addressing the dynamics of membrane proteins in
near physiological conditions, we employ molecular dynamics simulations
to rationalize the information gleaned in structural biophysics
experiment in mimetic environments, notably in detergents.Dehez, F.; Pebay-Peyroula, E.; Chipot, C. Binding of ADP in the mitochondrial ADP/ATP carrier is driven by an electrostatic funnel. J. Am. Chem. Soc. 2008, 130, 12725-12733. Zoonens, M.; Masscheleyn, S.; Comer, J.; Pebay-Peyroula, E.; Chipot, C.; Miroux, B.; Dehez, F. Mitochondrial uncoupling protein 2 in dodecylphosphocholine: Partly denatured and severely inactivated. J. Am. Chem. Soc. 2013, 135, 15174-15182. |
Molecular motors.
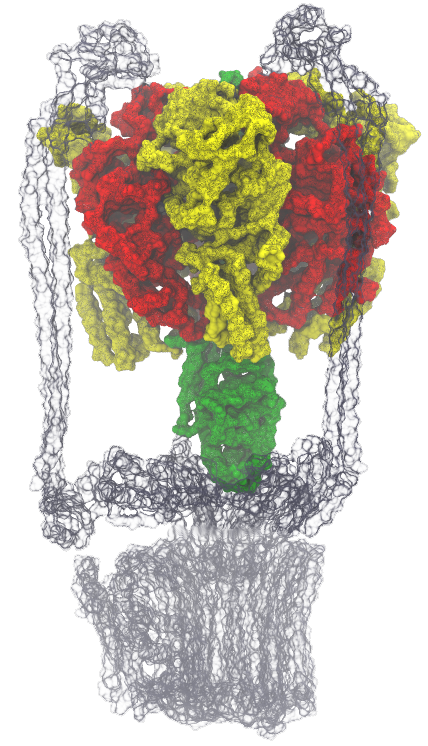 Molecular motors are nanoscale devices, which harness the free energy
from chemical reactions into mechanical work with minimal dissipation. Such
motors serve an important purpose in living organisms, driving
conformational transitions that regulate a variety of biological
processes, from RNA translocation, ATP synthesis and hydrolysis, to
cytoskeletal transport. Chemists have learned lessons taught by the
cell machinery and the principles of energy transduction in biological
motors to design and synthesize structurally simpler, yet functionally
targeted abiological devices. These devices have found applications in
various areas of molecular recognition, encompassing nanosensors and
transducers, which, in turn, can be employed in an automated platform
for the synthesis of small molecules, defining an area of frontier
research that was awarded the Nobel Prize in Chemistry in 2016. Our
research focuses on molecular motors at different scales, from small
abiological, cyclodextrin-based nanodevices to the large biological
complexes of the respiratory chain. Using molecular-dynamics
simulations, our effort aims at reconciling structural, biochemical,
thermodynamic and kinetic information to render a complete, detailed
picture of the processes at play. In addition, we focus on the spurious
mutations and oxidative stress that affects the efficiency of the
respiratory complexes, in connection with a variety of mitochondrial
diseases and aging.
Molecular motors are nanoscale devices, which harness the free energy
from chemical reactions into mechanical work with minimal dissipation. Such
motors serve an important purpose in living organisms, driving
conformational transitions that regulate a variety of biological
processes, from RNA translocation, ATP synthesis and hydrolysis, to
cytoskeletal transport. Chemists have learned lessons taught by the
cell machinery and the principles of energy transduction in biological
motors to design and synthesize structurally simpler, yet functionally
targeted abiological devices. These devices have found applications in
various areas of molecular recognition, encompassing nanosensors and
transducers, which, in turn, can be employed in an automated platform
for the synthesis of small molecules, defining an area of frontier
research that was awarded the Nobel Prize in Chemistry in 2016. Our
research focuses on molecular motors at different scales, from small
abiological, cyclodextrin-based nanodevices to the large biological
complexes of the respiratory chain. Using molecular-dynamics
simulations, our effort aims at reconciling structural, biochemical,
thermodynamic and kinetic information to render a complete, detailed
picture of the processes at play. In addition, we focus on the spurious
mutations and oxidative stress that affects the efficiency of the
respiratory complexes, in connection with a variety of mitochondrial
diseases and aging.Liu, P.; Shao, X.; Chipot, C.; Cai, W. The true nature of rotary movements in rotaxanes. Chem. Sci. 2016, 7, 457-462. Singharoy, A.; Chipot, C.; Moradi, M.; Schulten, K. Chemomechanical coupling in hexameric protein- protein interfaces harnesses energy within V-Type ATPases. J. Am. Chem. Soc. 2017, 139, 293-310. |

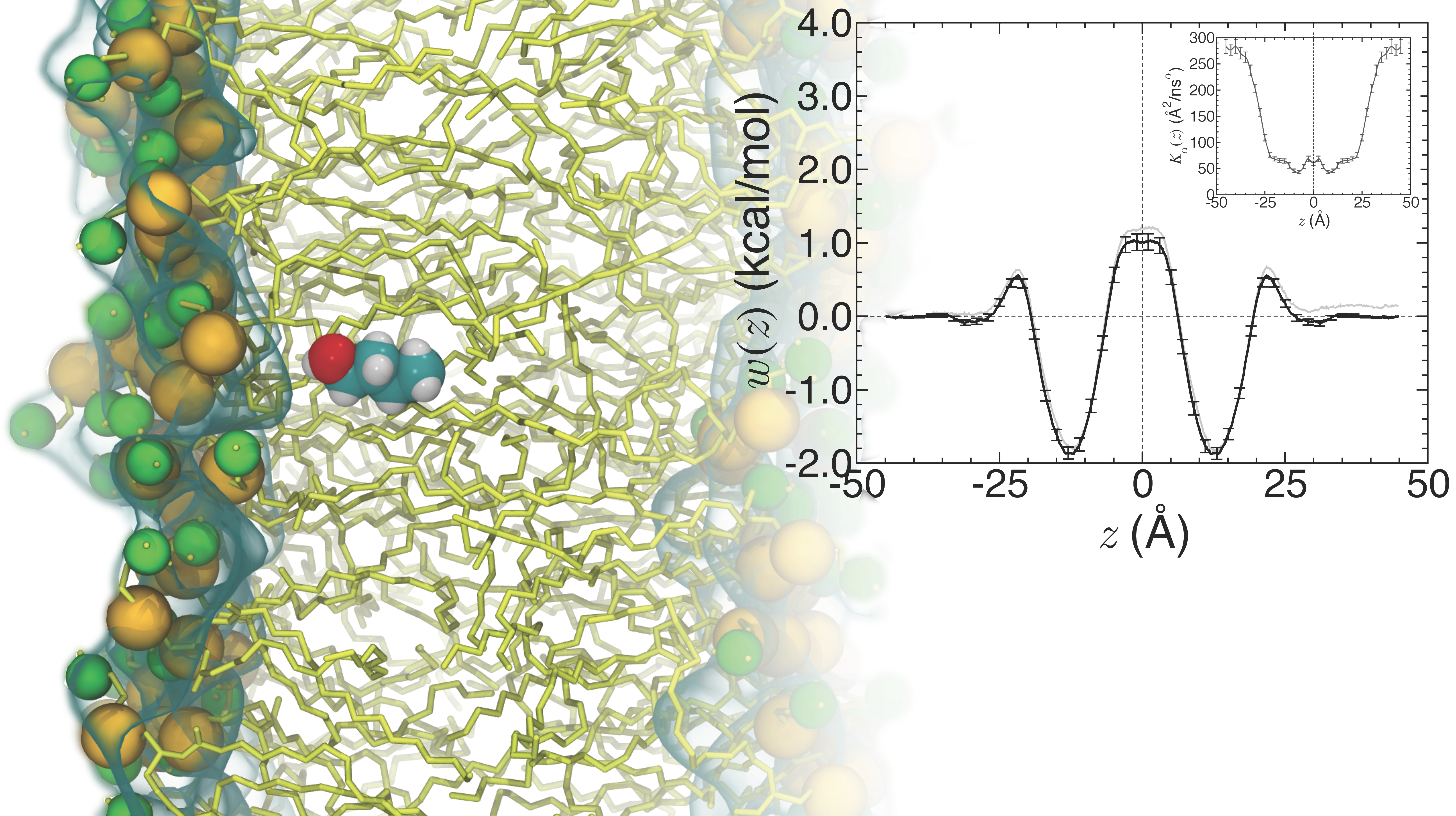 Concomitantly with the development of novel algorithms targeted at the
exploration of rare events, we are reconciling thermodynamics and
kinetics by means of Bayesian-inference schemes to build kinetic models
underlying biological phenomena. Exploiting gradient-based free-energy
calculations, our methodology supplies the position-dependent
diffusivity, from whence mean first-passage times and rate constants
can be inferred. This methodology has been recently extended to address
anomalous diffusion, whereby the mean-squared displacement along the
chosen coarse variable is no longer linear in time, turning to a
fractional Smoluchowski description.
Concomitantly with the development of novel algorithms targeted at the
exploration of rare events, we are reconciling thermodynamics and
kinetics by means of Bayesian-inference schemes to build kinetic models
underlying biological phenomena. Exploiting gradient-based free-energy
calculations, our methodology supplies the position-dependent
diffusivity, from whence mean first-passage times and rate constants
can be inferred. This methodology has been recently extended to address
anomalous diffusion, whereby the mean-squared displacement along the
chosen coarse variable is no longer linear in time, turning to a
fractional Smoluchowski description.

