Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
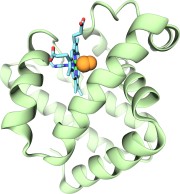
Many proteins interact with gas molecules such as oxygen to perform their
functions. In most cases, the gas molecules must reach sites buried deep
inside the proteins that bind the molecules, with no obvious way in.
Understanding how, for example, oxygen enters the protein, and mapping out
which pathways it takes has been a long-standing challenge. As reported
recently,
computational biologists, inspired by previous work on the hydrogenase
enzyme (see the September
2005 highlight), have developed a method, called implicit ligand
sampling, that maps the pathways taken by gas molecules inside proteins.
The mapping is determined by monitoring fluctuations of the protein,
surprisingly, in the absence of the gas molecules. The mapping method is
available in the most recent version of the program VMD used for structure
and sequence analysis of proteins. The
researchers applied the method to myoglobin, an oxygen-storing protein
present in muscle cells, and determined detailed three-dimensional maps of
oxygen and carbon monoxide pathways inside the protein (for more
information see our web page). While
some details of these pathways were already known from experiment, the
implicit ligand maps revealed a large number of new pathways and suggest
that oxygen enters myoglobin using many different entrance doors.