VMD 1.9.2
The Theoretical and Computational Biophysics Group is pleased to announce VMD version 1.9.2.VMD incorporates many new improvements for high quality rendering and export of molecular scenes, new analysis features, support for new molecular data file formats, and many performance improvements. Many new and updated structure building and analysis tools have been added in this release, easing the process of preparing, running, and analyzing biomolecular simulations. This release also contains many performance and efficiency improvements that are particiularly beneficial for modeling, visualizing, and analyzing very large structures with over 100 million atoms. VMD makes extensive use of multi-core processors and GPU acceleration to speed up computationally demanding analysis and visualization tasks including key structure and trajectory analysis features, interactive molecular dynamics, and high-quality ray tracing of molecular scenes.
- Updated Gromacs plugins for VMD 1.9.2
- VMD 1.9.2 Documentation, Release Notes, Tutorials
- Download VMD 1.9.2 for MacOS X, Unix, or Windows
- VMD 1.9.2 Development and Release History (large)
Major features included in VMD 1.9.2:
VMD 1.9.2 contains several updates for the Molecular Dynamics Flexible Fitting (MDFF) Method. The mdff plugin contains new options for setting up MDFF simulations with implicit solvent and the new MDFF method for low-resolution x-ray crystallography (xMDFF, recently detailed in this article). Complete details for setting up xMDFF simulations with this plugin can be found in the MDFF tutorial. Additionally, a new graphical user interface for the mdff plugin is now available in the Modeling section of the Extensions menu. This interface allows for easier setup of MDFF and xMDFF simulations, as well as running, connecting to, and analyzing interactive MD simulations. VMD 1.9.2 also contains new multi-core CPU and GPU-accelerated analysis capabilities for MDFF, as described in a recent article. The mdffi cc command can be used to quickly compute the cross correlation of a structure to a target density map much faster than previous methods. This speedup allows for the analysis of very large structures (millions of atoms) and very long (microsecond range) trajectories, and at a much finer level of detail than was previously feasible. The Timeline plugin uses this new fast cross correlation method to visually present the results of the analysis, making it easier than ever to determine the quality of fit for a MDFF simulation and to quickly identify poorly fitting regions of a structure that require further simulation.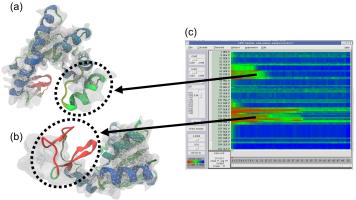 GPU-accelerated MDFF hybrid structure quality-of-fit
GPU-accelerated MDFF hybrid structure quality-of-fit
cross correlation analysis shown in the Timeline pluginVMD 1.9.2 contains many features for generating high quality renderings of molecular graphics and for creation of movies of both static structures and molecular dynamics simulation trajectories. VMD includes a new built-in GPU-accelerated version of the Tachyon ray tracing engine, using NVIDIA CUDA and OptiX to achieve performance levels up to six times faster than existing CPU versions of Tachyon. The new renderer is labelled "TachyonL-OptiX" in the VMD renderer window, and it supports the vast majority of Tachyon rendering features, with emphasis on ambient occlusion lighting, shadows, depth-of-field, high quality transparent surface rendering, and parallel movie rendering. The updated ViewChangeRender plugin provides an easy-to-use graphical interface for managing multiple VMD camera viewpoints and making movies that fly the camera between multiple viewpoints. The latest version of ViewChangeRender also supports parallel movie rendering on clusters and supercomputers.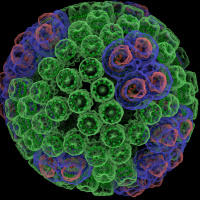
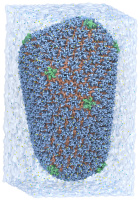
VMD molecular scenes with ambient occlusion lighting, shadows, and
angle-modulated transparency, rendered with the new GPU-accelerated
TachyonL-OptiX parallel ray tracing engine built-into VMD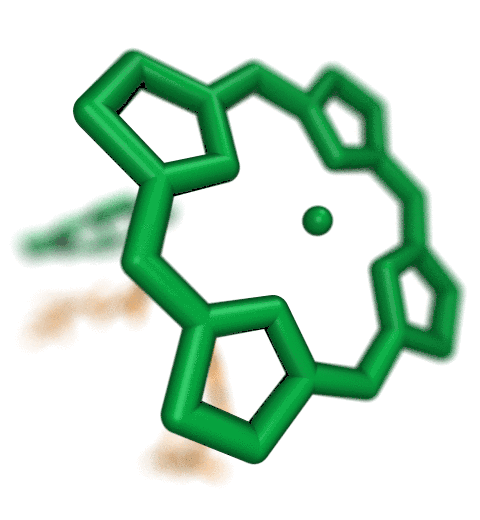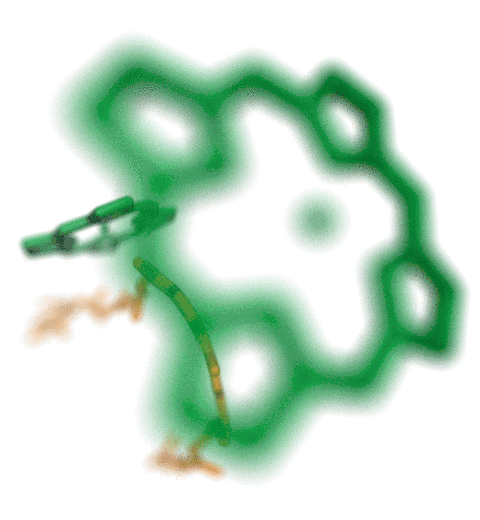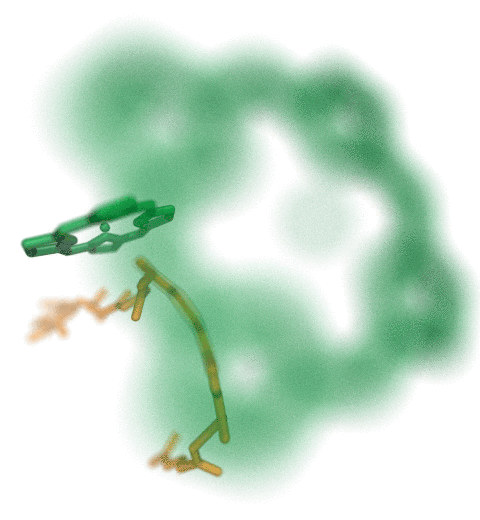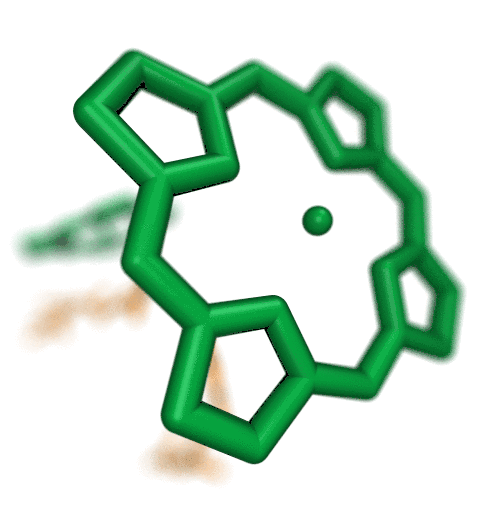

VMD molecular scenes with depth-of-field focal blur,
ambient occlusion lighting, and shadows, rendered with the
interactive TachyonL-OptiX GPU ray tracing engine built-into VMDThe speed of the new GPU-accelerated ray tracing feature allows VMD users to interactively visualize moderate size molecular complexes containing a few million atoms with shadows, ambient occlusion, and on fast GPUs, even the depth-of-field focal blur effect can be rendered in real-time. The new interactive ray tracing feature uses a progressive refinement approach to allow users to manipulate the view using the mouse, and once the mouse is released, the interactive ray tracing engine refines the image with monte carlo sampling of lighting, focal blur, and so on. The interactive ray tracing engine allows multiple views to be saved at any time, as well as the default behavior of saving the final image shown when the user exits the viewer. Future versions of VMD will more closely integrate interactive ray tracing, enabling interactive ray tracing in the main VMD window, as an alternative to the traditional OpenGL-based visualization engine in VMD.
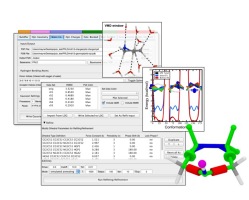
The Force Field Toolkit (ffTK) plugin provides a comprehensive toolset for the development of CHARMM-compatible (e.g., CGenFF) force field parameters, including charges, bonds, angles, and dihedrals. VMD 1.9.2 includes a vastly improved version of ffTK boasting many new features: a faster charge optimization routine; optimization of bond and angle parameters; projection of missing parameters onto molecular structure; an interactive display for analyzing complex dihedral potential energy surfaces within the context of the molecular structure. All tools are accessed through the provided GUI, which organizes parameterization tasks into a clear and modular workflow. Several embedded plotting utilities are included to assess parameter performance on-the-fly, provide users with quantitative feedback useful in tuning optimization routines, and allows easy export of data for use in publication and presentation figures. Documentation and screencast tutorials are available on the plugin website.As reported in several publications, VMD uses GPUs to accelerate many of the most computationally demanding visualization and analysis features of interest to biomedical researchers. VMD 1.9.2 advances these capabilities further, adding an improved GPU-accelerated implementation of the QuickSurf molecular surface representation that greatly reduces amount of GPU memory required when displaying large biomolecular complexes, and provides a roughly 2x performance increase compared to VMD 1.9.1. The QuickSurf representation enables smooth interactive animation of moderate sized biomolecular complexes consisting of a few hundred thousand to one million atoms, and interactive display of molecular surfaces for multi-million atom complexes such as virus capsids. The range of acceleration provided by GPUs depends on the capabilities of the specific GPU device(s) installed, and the details of the calculation. Typical (approximate) acceleration factors for the algorithms in VMD comparing a single high-end GPU vs. a single quad-core CPU are: molecular surface display 2.5x to 11x, electrostatics 5x to 11x, implicit ligand sampling 4x to 7.5x, calculation of radial distribution functions 7.5x to 22x, molecular orbital calculation 10x to 30x. Details on making best use of the GPU acceleration capabilities in VMD are provided here.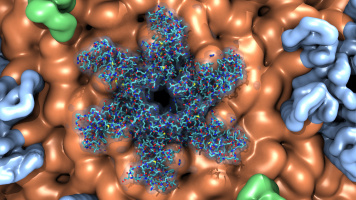
Fast "QuickSurf" multi-resolution molecular surface
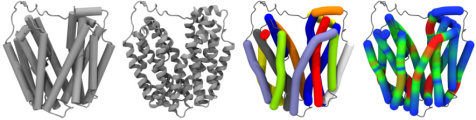
calculation and display with CUDA GPU accelerationBendix is a user-friendly plugin for VMD, that makes it easier to characterize proteins. It allows VMD to calculate and visualize both dynamic and static helix geometry, and abstracts helices without sacrificing conformation. Moreover it accepts both coarse-grained and atomistic proteins. Bendix displays protein helices as cylinders that follow the helix axis. This captures conformational information that is lost by classical, straight cylinders. Coarse-grained proteins also benefit from beta-sheet and backbone display. Helix geometry can be analysed both qualitatively, using the revealing heatmap colour-coding, and quantitatively, where data is 2D or 3D graphed, with the option to export for viewing with common graphing packages. With its multiple settings, Bendix is easily tailored. Bendix integrates with the rest of the built-in VMD graphical representations. Please view the slideshows and the Gallery and Tutorial sections of the Bendix home page for examples of its use.
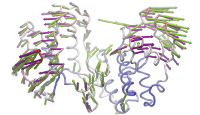
ANM modes for p38 MAPK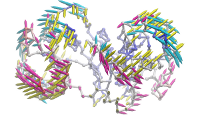
ANM vs. PCA modes for p38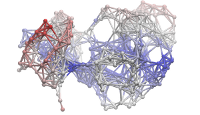
p38 network modelThe Normal Mode Wizard (NMWiz) plugin allows visual comparative analysis normal mode data.The updated Normal Mode Wizard (NMWiz) plugin permits visual comparative analysis of normal mode data. Normal modes may come from normal mode analysis (NMA) of elastic network models (ENMs), e.g. ANM/GNM, force-field based all-atom NMA, principal component analysis (PCA) of experimental structure ensembles, or essential dynamics (EDA) of molecular dynamics trajectories. NMWiz can draw normal mode arrows, make animations (conformations along a normal mode), plot square-fluctuations (labeling and highlighting of residues), and compare pairs of structures drawing deformation arrows.
The updated remote control feature of VMD 1.9.2 provides the ability for multiple users to collaboratively control a VMD session from mobile phones and tablets. The remote control app allows mobile devices to manipulate the VMD molecular display, moving, rotating, and scaling the displayed molecular structure. Several user-defined remote control buttons can be used to trigger VMD scripts, drive presentations using an updated version of the ViewMaster plugin. The latest version of the remote control Android app includes a greatly simplified startup process, the ability to take image "snapshots" of the VMD OpenGL window and then Share them with others via Android's "Share" mechanism, and improved configurability.
The NetworkView plugin provides a graphical interface for the study of allostery and signalling using network models, allowing networks to be mapped onto molecular structures loaded in VMD. New in VMD 1.9.2 is improved support for very large (>1 gigabyte) data files. Users can now scale suboptimal path counts, and drawing performance has been improved.