As
reported in several publications,
VMD uses GPUs to accelerate many of the most computationally demanding
visualization and analysis features of interest to biomedical researchers.
VMD 1.9.1 advances these capabilities further, adding a GPU-accelerated
implementation of the new QuickSurf molecular surface representation,
enabling smooth interactive animation of moderate sized biomolecular
complexes consisting of a few hundred thousand to one million atoms,
and interactive display of molecular surfaces for multi-million atom
complexes, e.g. large virus capsids. The GPU-accelerated QuickSurf
representation in VMD achieves performance orders of magnitude faster than
the conventional Surf and MSMS representations, and makes VMD the first
molecular visualization tool capable of achieving smooth animations
of surface representations for systems of up to one million atoms.
The range of acceleration provided by GPUs depends on the
capabilities of the specific GPU device(s) installed, and the details
of the calculation. Typical acceleration factors for the algorithms
in VMD on a single high-end GPU are:
molecular surface display 10x to 45x,
electrostatics 22x to 44x,
implicit ligand sampling 20x to 30x,
calculation of radial distribution functions 30x to 90x,
molecular orbital calculation 100x to 120x.
Details on making best use of the GPU acceleration capabilities
in VMD are
provided here.
VMD 1.9.1 contains many features for generating
high quality renderings of molecular graphics and for creation
of movies of both static structures and molecular dynamics simulation
trajectories. The updated
ViewChangeRender
plugin provides an easy-to-use graphical interface for managing multiple
VMD camera viewpoints and making movies that fly the camera between
multiple viewpoints. The VMD Wavefront OBJ export feature has been
extensively tested with the most recent versions of Autodesk's
Maya, and
3ds-Max professional
rendering tools. VMD also supports export to the RenderMan .rib file
format used by
Pixar's RenderMan,
the open source renderer
Aqsis, and
many other animation and rendering tools.
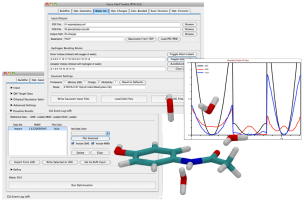
New Force Field Toolkit plugin assists with
development of CHARMM-compatible parameters

VMD remote control app for mobile
Android phones and tablets
The newly added
NetworkView
plugin provides a graphical interface for the study of allostery and
signalling using network models, allowing networks to be mapped onto
molecular structures loaded in VMD.
The new
Force Field Toolkit
(FFTK) plugin provides tools that aid users in the
development of CHARMM-compatible forcefield parameters,
including charges, bonds, angles, and dihedrals.
These tools are accessed through the provided GUI, which greatly
simplifies the setup and analysis of the underlying calculations. Notable
features include assisted generation of Gaussian input files and parsing of the
resulting log files to obtain QM target data, access to downhill and simulated
annealing optimization routines, and interactive fitting of dihedral parameters
via embedded data plotting tools.
The updated
Timeline plugin
provides an interface for viewing temporally changing per-residue
attributes of a molecular structure. It can also display temporally changing
attributes of a set of VMD selections, for example a set of all the
salt-bridge pairs observed in a trajectory.
The controls allow selection of the molecule, or part of the molecule,
used for the calculation.
The graphical display of residues and timesteps can be scrolled and
zoomed as necessary to see results for large structures and long trajectories.
The latest version significantly improves the display of large
structures and long timescale trajectories.
The newly added
PropKa
interface for graphical analysis of pH-dependent properties of
proteins using the PROPKA package.
The new
NMWiz plugin
provides a graphical interface for normal mode analysis.
The
RMSD Trajectory Tool has
been updated with many new features including swapping of equivalent
atoms, and calculation of average, std. deviation, and other useful statistics.
The new
RMSD Visualizer plugin
allowed RMSD and RMSF values to be computed for specified atom selections
and plotted using the new
HeatMapper
3-D heat map generation tool and the
Multiplot plugin.
A new
remote control
feature of VMD 1.9.1 provides the ability to control a VMD session
from mobile phones and wireless tablet devices. The remote control app
allows mobile devices to manipulate the VMD molecular display, moving,
rotating, and scaling the displayed molecular structure. Several user-defined
remote control buttons can be used to trigger VMD scripts, drive presentations
using an updated version of the
ViewMaster plugin.
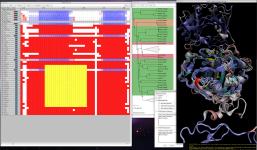
MultiSeq analyzes large datasets efficiently
This release of VMD includes the newly revised
MultiSeq plugin.
MultiSeq includes updated support for
MAFFT
for multiple sequence alignments. The latest version improves support
for ClustalW and MAFFT alignments for sequences containing non-standard or
unrecognized residues.
Non-redundant set calculations can now be performed for a
set of user-selected sequences.
Major efforts have been directed toward improving the ability of MultiSeq
to handle large data sets, and the new MultiSeq is capable of loading
and analyzing 100,000 sequences on a typical desktop machine.
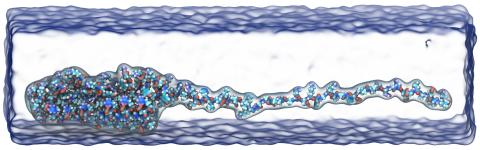
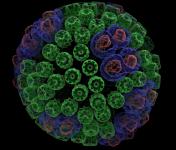
 VMD molecular scenes with ambient occlusion lighting, shadows,
VMD molecular scenes with ambient occlusion lighting, shadows, New Force Field Toolkit plugin assists with
New Force Field Toolkit plugin assists with VMD remote control app for mobile
VMD remote control app for mobile



