Various Systems
As the NIH Resource for Resource for Macromolecular Modeling and Bioinformatics,
the group invites collaborations with biomedical and biomaterials researchers
on new and challenging projects. Projects of this kind include the design
of novel biomaterials and proteins, and simulations of very large scale systems
(eg ribosome), chaperone proteins (eg GroEL, GroES), and molecular
motors.
The group favors studies which can benefit from our expertise in large scale
systems, physical analysis, molecular electronics, and Interactive Molecular
Dynamics.

|
Through modelling and quantum
chemical studies, the group is supporting the design of novel
proteins in collaboration with M. Sarikaya, U. Washington, Seattle.
It is hypothesized that
electrostatic interactions between the polar residues of this genetically
engineered polypeptide and
the gold surface allow stronger adsorbtion onto the {111} surface than to
other Au crystal faces, thus influencing the crystalization of gold
in the presence of this polypeptide.
|
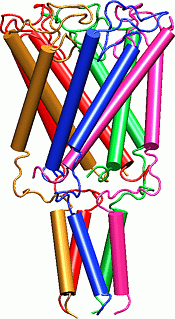
| Mechanosensitive channels (MscL) open a conductance pore in cell membranes
in response to mechanical stress. Molecular dynamics simulations of the
channel-membrane system can provide a description of the mechanical
properties of this channel. We plan to simulate a MscL-POPC membrane system
at constant surface tension, corresponding to the closed-to-open transition
tention as determined experimentally.
| 
|

|
The discovery of the crystal structure of the kinesin motor domain has made it possible to
study kinesin's dynamics by computer simulations. To model pH- and nucleotide-dependent changes in the kinesin structure, we carried
out conformational searches by simulated annealing. The conformational differences of the ATP-bound protein relative to the ADP-bound state can be attributed to a force-producing power stroke.
|






