Cytochrome bc1 Complex
Quinol binding at the Qo-site
The bc1 complex is an important enzyme involved in the conversion of energy into a proton gradient across the cellular membrane in photosynthetic bacteria, and cellular respiratory systems in higher organisms. The resulting gradient is subsequently used in the synthesis of ATP and the production rate of the last depends on the effectiveness of the bc1 complex. The reaction mechanism, namely the Q-cycle, consists of a series of charge transfer reactions between cofactors, such as quinones and quinols, and different bc1 complex subunits.
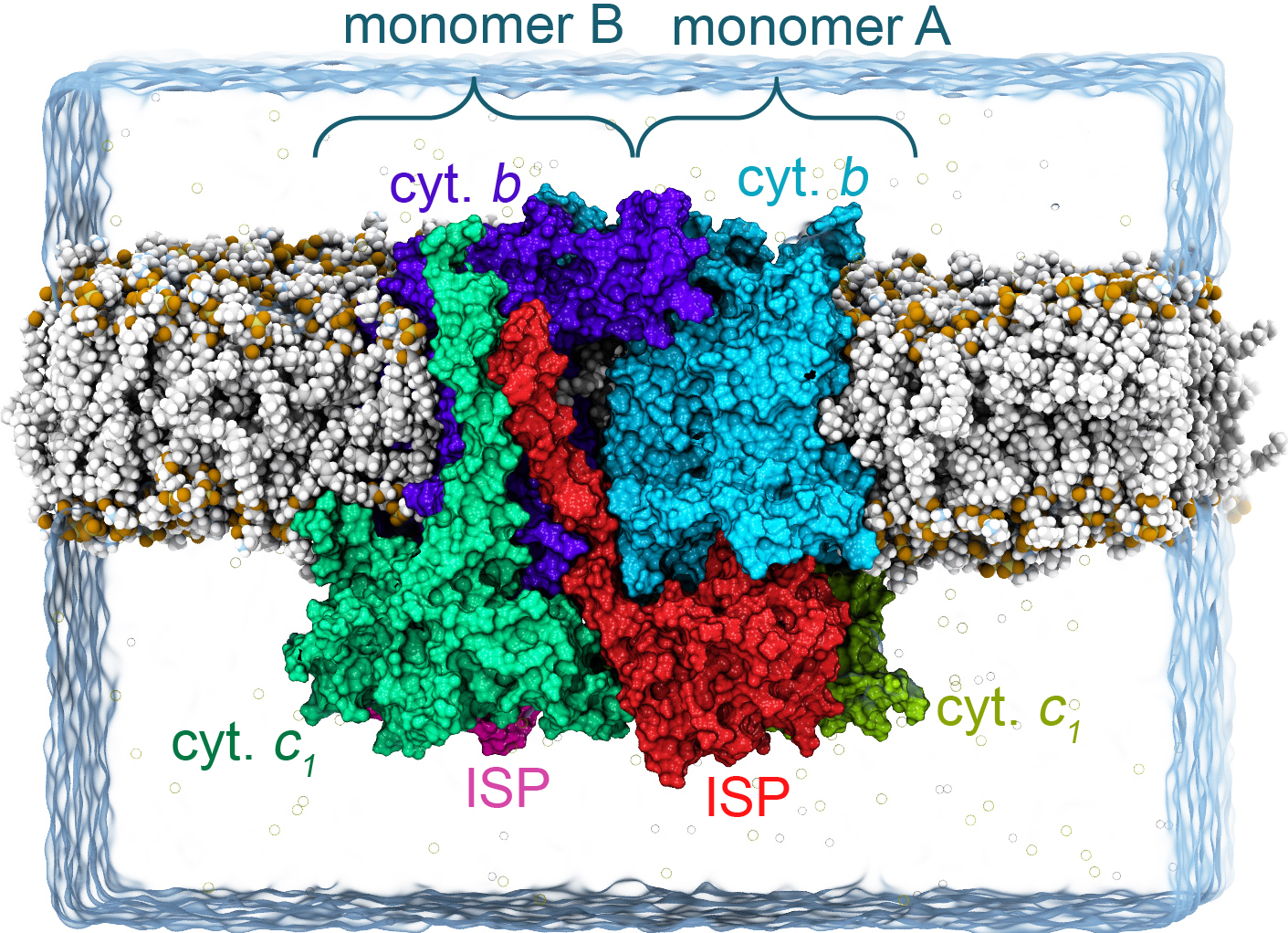
Fig. 1. Simulation box of the bc1 complex from Rhodobacter Capsulatus. The bc1 complex consists of a dimeric arrange of protein subunits that contain heme groups, involved in the electron transfer processes between cofactors.
In the primary step, a quinol molecule binds the bc1 complex at the Qo-site. There, two electron are transferred from quinol and to heme groups from cyt. b and cyt. c1 protein subunits. Each electron transfer is coupled to a proton transfer, and each pair of proton-coupled electron transfers (PCET) leads to the transformation of one quinol into a quinone. As a result of such transformation, one proton is pumped to the positive side of the membrane, creating the proton gradient.
Despite important discoveries made throughout the last decades, the reaction mechanism of the bc1 complex has not been elucidated and molecular detail of both binding and charge transfer reactions are lacking of an accurate description. Furthermore, controversies around the protonation states of the protein amino acids involved in the docking and following reactions, have been raised. By using powerful tools such as molecular dynamics simulations and ab-initio quantum chemical calculations, we investigate the quinol binding and subsequent charge transfer reactions at atomistic level.
By comparing two different binding schemes of the quinol to the Qo-site, a recent study revealed novel configurations of the key side groups at the binding site, such as H156, Y147 and E295, that stabilize the reaction complex. These key amino acids also provide an optimal configuration prior to the charge transfer reactions between quinol and iron-sulfur cluster of the iron-sulfur protein (ISP). Re-arrangements in the E295 and Y147 side chains were observed in all our simulations, showing intermediate bridging hydrogen bonding between quinol and E295, not observed before.
Unveiling the reaction mechanism of the bc1 complex has important implications in the understanding of energy conversion in photosynthesis and respiration. The bc1 complex malfunctions in the respiratory chain are central to a large number of diseases associated with aging, arthritis, cancer, heart disease, diabetes and obesity. Likewise, the high efficiency of energy conversion in the bc1 complex during photosynthesis, makes it of outstanding relevance for optimal energy conversion research.
Recognition and docking of the Cytochrome c2 to the bc1 complex
Small diffusible redox proteins facilitate electron transfer in respiration and photosynthesis by alternately binding to integral membrane proteins in a redox state selective manner and exchanging electrons. Given the redox proteins' ubiquitous role in sustaining biological energy conversion processes, their interaction with their integral membrane counterparts has been studied, in the case of cytochrome c2 (cyt. c2) with the bc1 complex, for over 30 years, in particular, pertaining to the binding-site recognition, binding and unbinding of cyt. c2 that often present major kinetic bottlenecks. Evolutionary data suggests that these proteins might invoke extremely precise molecular-scale functional designs to optimize the kinetic bottlenecks across several species; yet an unified mechanism of cyt. c2 turnover remains elusive as of now. The present study provides a first all-atom description of the cyt. c2 - docked bc1 complex in Rhodobacter sphaeroides in terms of an ensemble of favorable docking conformations.
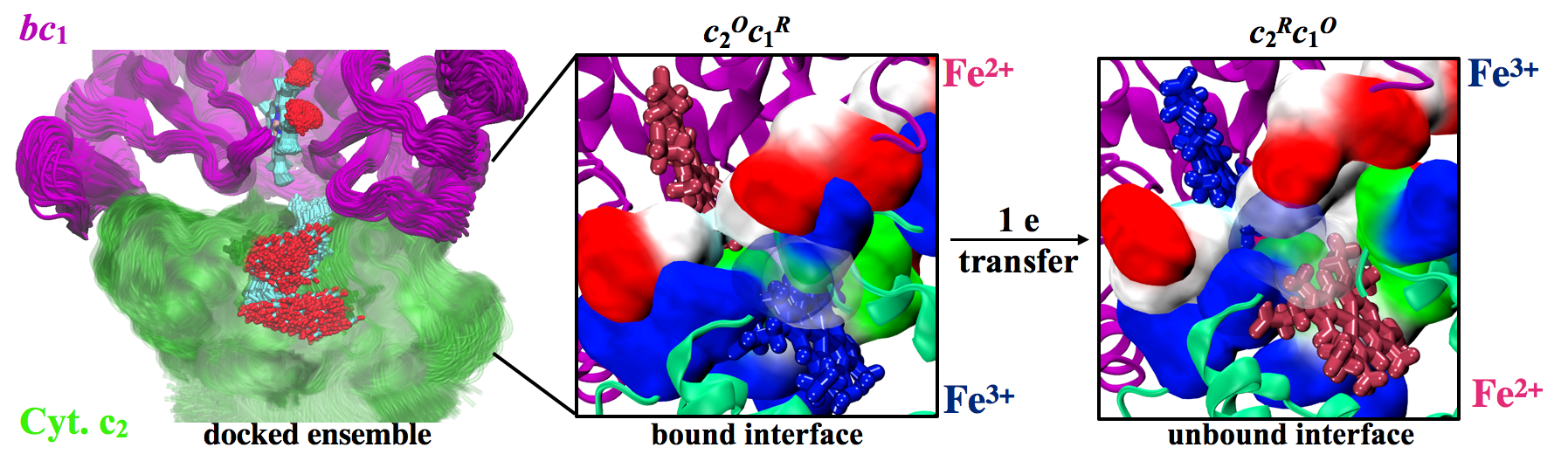
Molecular dynamics (MD) simulations, performed for this purpose for different redox states of the participant heme groups, reveal an intricate series of conformational changes that allow cyt. c2 to recognize the cyt. c1 subunit of the bc1 complex and depending upon the redox states of the associated hemes to bind or to unbind. These conformational changes are perfectly poised to tune kinetics and thermodynamics of the cyt. c2 bindability to the bc1 complex every time an electron is transferred from cyt. c1 to cyt. c2: prior to the electron transfer, an oxidized cyt. c2 samples multiple docking poses before binding to the reduced cyt. c1 via a unique half-ring arrangement of complementary residues; a direct heme-to-heme electron transfer enhances mobility of the positively charged residues on cyt. c2 surface inducing thereafter a water-mediated conformational switch that reshapes the cyt. c2 - cyt. c1 interface and promotes cyt. c2 unbinding. The exemplary precise functional design revealed by the cyt. c2 binding/unbinding pathways appear to permit single electron transport across the broader cyt. c family of proteins. Finally, the unified modus operandi of cyt. c mediated electron transport discovered here is employed to successfully resolve long-standing discrepancies between structural data and functional studies.
Proton-coupled electron transfer at the bc1 complex
The charge tranfer reactions taking place upon quinol binding at the Qo-site of the bc1 complex were investigated. The combination of quantum chemistry calculations and all-atom molecular dynamics simulations allowed to unveil the nature of the proton-coupled electron transfer reaction between the quinol and the ISP subunit of the bc1 complex. The video shows the highest occupied molecular orbital (HOMO), as blue and orange surfaces, and the position of the transferring proton, as a magenta sphere, for a set of snapshots along the reaction energy profile. The proton and the electron are delocalized between their donor and acceptors at the transition state.
TCB Publications
Mechanism of the primary charge transfer reaction in the cytochrome bc1 complex. Angela M. Barragan, Klaus Schulten, and Ilia A. Solov'yov. Journal of Physical Chemistry B, 2016. DOI: 10.1021/acs.jpcb.6b07394
Binding site recognition and docking dynamics of a single electron transport protein: Cytochrome c2. Abhishek Singharoy, Angela M. Barragan, Sundarapandian Thangapandian, Emad Tajkhorshid and Klaus Schulten. Journal of the American Chemical Society, 138:12077-12089, 2016.
Identification of ubiquinol binding motifs at the Qo-site of the cytochrome bc1 complex. Angela M. Barragan, Anthony R. Crofts, Klaus Schulten, and Ilia A. Solov'yov. Journal of Physical Chemistry B, 119:433-447, 2015.
Steered molecular dynamics simulation of the Rieske subunit motion in the cytochrome bc1 complex. Sergei Izrailev, Antony R. Crofts, Edward A. Berry, and Klaus Schulten. Biophysical Journal, 77:1753--1768, 1999.
Investigators
maintained by Angela M. Barragan



