Recognition and Assembly
Macromolecules of the living cells rarely exist and function alone.
As a rule, they act as parts of complex biomolecular assemblies, which may
involve several protein units, DNA loops, lipids, various ligands,
etc. The Theoretical Biophysics Group focuses on structure
prediction of biomolecular assemblies and studies molecular recognition
and assembly.
The employed research methods include molecular dynamics, molecular
replacement, and multi-resolution modeling.
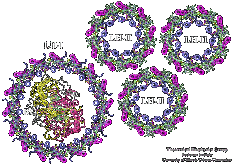
| Photosynthetic units (PSUs) of living organisms are complex assemblies
of several multi-protein aggregates. We constructed the structure of the
PSU of purple bacteria. The components involved were the crystal
structure of the bacterial photosynthetic reaction center surrounded by
the model structure of the light-harvesting complex I and several
structures of the light-harvesting
complex II, solved in a collaborative project with X-ray
crystallographers. The constructed structure is used to study the light
energy transfer driving the bacterial photosynthesis. |
 |
 |
The structure of a 160-lipid bilayer surrounded by two apoA-I lipoproteins has been built on the basis of available experimental data and refined in the course of molecular dynamics simulations. |
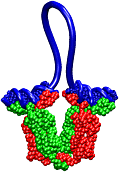
| A coarse-grained method of DNA modeling has been developed as a stepping-stone towards a multi-resolution approach to biomolecular modeling. The DNA structure is obtained after numerically solving a Kirchhoff system of equations, augmented with electrostatic and steric repulsion force terms. The method has been applied to restore the missing structure of a DNA loop clamped by the lac repressor. |
 |
 |
A chromosomal protein HMG-D has been docked to several conceivable DNA segments in order to predict the structure of a complex between the two macromolecules. The data obtained in the course of a molecular dynamics simulation allowed us to select one DNA-protein complex as a likely candidate for the correct structure. The prediction turned to be in a good agreement with the recently published crystal structures of this and related protein-DNA complexes. |
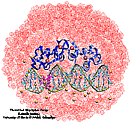
| Nuclear hormone receptors are cellular regulators which activate the
transcription of specific genes in response to the binding of nuclear hormones. We studied the specificity of DNA recognition by the estrogen receptor protein. The role of water molecules at the protein-DNA interface and changes in the DNA structure between specific and non-specific binding were monitored and analyzed. |
 |
 |
DNA hydration plays an important role in DNA dynamics and protein-DNA interaction. We have computationally studied the effect of DNA analogue bases on DNA hydration, to complement the experiments of our collaborators. The results suggest that the analogue provides a good mimic of water molecules, specifically bound to DNA. |
| The enzyme called phospholipase A2 plays an important role in cell signalling. The enzyme cleaves phospholipids of cellular membranes, producing the arachidonic acid, which serves as a precursor of signalling molecules on several cell signalling pathways. In order to elucidate the mechanism of the enzyme-membrane interaction, we docked the crystal structure of the phospholipase A2 to the lipid membrane and studied the resulting protein-membrane complex in series of molecular dynamics simulations. |
 |
 |
Actin protein units form filaments in the cells in order to manage the cellular shape changes, cell locomotion and chemotactic migration. The mechanical properties of the filaments substantially depend on the ATP hydrolysis by actin. Using molecular dynamics, we studied the ATP hydrolysis and the subsequent release of the inorganic phosphate.
|











