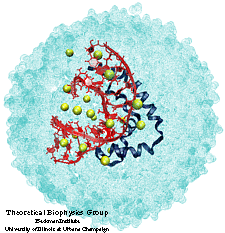
HMG-D protein - DNA complex
HMG-D is a sequence-non-specific DNA binding protein of Drosophila melanogaster. The protein belongs to a diverse family of HMG proteins. A unique feature of the family is that it includes both proteins interacting with DNA sequence-specifically and proteins interacting with DNA sequence-non-specifically. Although the proteins recognize DNA in two different ways, they have a homologous DNA binding domain called HMG-box. The structure of the domain was obtained in 1994 and presented a novel DNA binding motif. Another interesting feature of the HMG proteins is their architecture specificity, that is an increased affinity for distorted regions of DNA, which can be even larger than the natural affinity of the sequence-specific HMG proteins for their recognition sites.
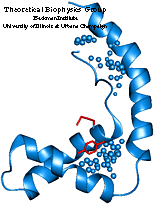
HMG-D is a chromosomal protein. It was suggested that it functions as a DNA chaperon facilitating the packing of DNA into nucleosomes (Ner et al., 1994). The structure of the HMG-box of HMG-D has been obtained by NMR (Jones et al., 1994), but the structure of its complex with DNA has not been experimentally obtained yet. By means of molecular modelling, we predicted the structure of the complex between HMG-D and DNA.
Recently, the structures of complexes of two sequence-specific proteins of the HMG-1/2 family with DNA were solved. They confirmed the previous results, but exhibited some differences, especially in the location of the proteins' C-termini. Thus, it would be of great interest to determine the structure of a non-sequence-specific member of the family complexed with DNA. Our goal is to predict the structure of the HMG box of the DNA chaperon HMG-D in a complex with DNA, which we hope will improve our general understanding of DNA packing into nucleosomes.
As a tool for theoretical prediction of the structure, we have chosen molecular dynamics (MD) simulations which our group has used already as a powerful method for bridging experimental and theoretical studies of protein-DNA complexes. The simulation of highly charged systems like protein--DNA complexes requires the use of an aqueous environment and of full Coulomb interactions. This makes such simulations very computer-time intensive.
To prove the validity of the predicted structure we seek to reproduce the results of mutagenesis studies of HMG-1/2-proteins. We will use MD to estimate free energy changes due to mutations in the protein. This work will be carried out in collaboration with the experimental laboratory of M. Churchill at the U. of Colorado.
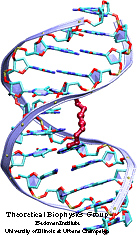
We have built a set of trial complexes between the HMG box of HMG-D and DNA, based on the NMR structure of the protein. To approximate the structure of DNA we used either a 12-bp DNA segment, pre-bent by a disulfide cross-link, or a 12-bp DNA segment from a complex with the TATA-binding protein; both DNA segments were bent to the degree necessary to interact with HMG-D. DNA was positioned on the protein on the basis of NMR, footprinting, mutagenesis and other experimental data.
After short energy minimizations, the trial complexes were placed into a 35 angstron-radius sphere of water molecules. After a 15 ps equilibration of the waters around the fixed complex, Na+ ions were placed in the solvent at the minima of the electrostatic energy to model physiological ionic conditions. The resulting systems were equilibrated for 30 to 50 ps, followed by 150 ps runs of free MD simulation. We determined various structural and dynamical parameters, e.g., the stability of the rms deviation, the quality of Met13 intercalation (by distance to nucleotide bases), the DNA geometry. We have recognized a trial complex of the HMG box with TATA--DNA as best stabilized, most consistent with the experimental data, and, thus, most probably corresponding to the real structure of the complex.
Additional simulations are planned to further sample the configurational space of the predicted structure, assuring it's stability. Recently, the structures of DNA with sequence-specific HMG-1/2 proteins became available, and we plan to repeat our computational experiment with these new structures taken as the initial guess, adapting their sequence to the HMG-D sequence. The refinement will be carried out to verify our predicted structure and to demonstrate the validity of our method.