Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
molecular dynamics flexible fitting (MDFF)
to ..." />
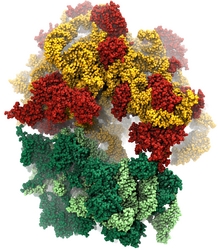

image size:
347.6KB
made with VMD
Water is the essential solvent that shapes protein structure and function,
but for researchers using
molecular dynamics flexible fitting (MDFF)
to fit large biomolecular models to data from cryo-electron microscopy, such as fitting the classical ribosome into the ratcheted map, it was a mixed blessing.
Since the network of hydrogen bonds that gives water its unique properties
must rearrange as the solute moves, water molecules not only increase
the size of the simulation but slow the fitting process.
Leaving water out completely was a common practice, relying on the
MDFF fitting potential to prevent the dehydrated protein from shriveling.
The 2.8 release
of NAMD
provides a better option: a new implementation of the
generalized Born implicit solvent model that scales to thousands of cores
for large biomolecular aggregates thanks to NAMD's unique parallel structure
and measurement-based load balancing system.
By eliminating explicit water molecules from the simulation, an
implicit solvent model helps shape protein structure while adapting
immediately to new conformations.
With this best-of-both-worlds option now available in NAMD,
biomedical researchers using MDFF need fear water no longer.



