Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
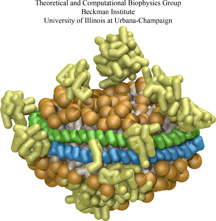
We all know sushi rolls, but just to be sure here is an easy definition: a wrapper encircles rice which holds a precious bit of fish. To make a sushi role is an
art and the same holds true for molecular sushi that is made of two lipoproteins as wrapper, lipids as rice, and membrane proteins as filling. Sushi rolls are for
eating. Molecular sushi roles are for holding membrane proteins in place for physical analysis; they actually come only in sliced form, one disc at a time. Due to
their size, the discs are called nanodiscs. Since membrane proteins are notoriously difficult to study experimentally due to their need to be in a "native"
membrane environment, nanodiscs are a great tool, furnishing a membrane environment that has been used to embed a variety of membrane proteins for biochemical
assay, including cytochrome P450's, rhodopsin, bacterial chemoreceptors, blood clotting factors, and translocation proteins. Unfortunately, it is difficult to make
either real or molecular sushi rolls (nanodiscs). In either case one needs to lay down the ingredients first. In the case of nanodiscs, one starts from the raw
ingredients which are solubilized by the detergent cholate. Removing the detergent allows the nanodiscs to self-assemble. However, the assembly process is
difficult to quantify experimentally, thus researchers rather studied the disassembly process, i.e., how detergent disassembles preformed nanodiscs. One can watch a sushi
chef make rolls, but watching the disassembly and assembly of nanodiscs is harder due the the small size. Fortunately, a computer can image the process. In a recent publication, nanodisc disassembly through the addition of increasing
concentrations of cholate was monitored through computer simulations using NAMD and verified through
experimental small-angle X-ray scattering. The study showed how cholate molecules insert themselves at the interface between the lipids and lipoproteins towards
complete disassembly. The simulations employed a new method called residue-based coarse-graining. For
more information, see our webpage on nanodiscs.