Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
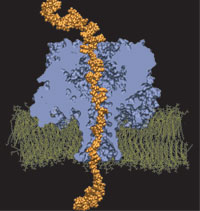
For the sequence of DNA, the genetic instructions of cells, to be read, the double helix of DNA is split open, exposing single DNA strands to DNA binding proteins. Once bound to DNA, the proteins, in carrying out their functions, will crawl along the DNA strand in one of two directions, towards DNA's 3' or 5' end. A recent study of DNA discovered a surprising property of single DNA strands that seems to explain how DNA binding proteins recognize the right direction on DNA strands. By measuring the translocation of DNA through alpha-hemolysin, a membrane protein with a narrow pore, researches discovered that directed single stranded DNA moves much faster when entering the pore 3' end first, rather than 5' end first. The underlying mechanism of this directionality was discovered through molecular dynamics simulations using NAMD and VMD. The simulations revealed that, in a narrow pore, DNA bases tilt collectively towards the 5' end, transforming a wide space directionless DNA brush into a tight space DNA ratchet. The 360,000-atom MD simulation did not only reveal how the DNA bases align and move faster in the "smooth" direction, but did also predict how the directional DNA movement can be discerned by means of direction-sensitive ionic currents through the channel blocked by translocating DNA strands. More details about this study can be found here.