Highlights of our Work
2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
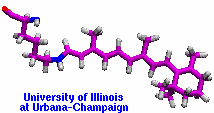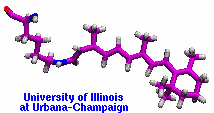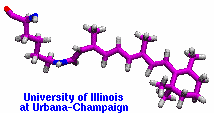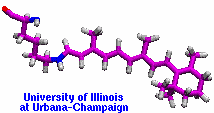
image size:
77.8KB
made with VMD
Retinal proteins are photoreceptors found in many living organisms. They possess a common chromophore, retinal, that upon absorption of light isomerizes and, thereby, triggers biological functions ranging from light energy conversion to phototaxis and vision. The photoisomerization of retinal is extremely fast, highly selective inside the protein matrix, and characterized through optimal sensitivity to incoming light. Photoisomerization takes place while the molecule (retinal) is in its short-lived electronically excited state when internal forces differ strongly from those in the prevalent ground state. Simulation of photoisomerization inside a protein has been a key goal, but unattainable so far since the forces of excited state molecules, largely unknown, need to be described by so-called ab initio quantum mechanical calculations and updated extremely frequently. A recent report describes that this goal has finally been achieved for retinal inside bacteriorhodopsin, a microbial retinal protein that functions as a light-driven proton pump, i.e., a pump driven by photoisomerization and thermal re-isomerization. The description unveils in complete detail the photoisomerization process and the essential role of the protein for the characteristic kinetics and high selectivity of the biomolecular reaction. Supported by comparison with dynamic spectral modulations observed in femtosecond spectroscopy, the results identify the principal molecular motion during photoisomerization that initiate proton pumping. Read more about the project.



