Till Rudack

My general areas of interest are computational chemistry and molecular structural biology. I am especially interested in investigating biological mechanisms in order to develop a fundamental understanding of cellular processes. I enjoy working in the interdisciplinary field of biophysical protein science in international teams and collaborative projects. In my graduate work, I studied at the interface between physics, biology, and chemistry as well as at the interface between theory and experiment.
During my doctorate I developed methods to combine results from biomolecular simulations with results from time-resolved FTIR spectroscopic measurements. With these methods I was able to investigate the small GTPase Ras from the atomic to the molecular level by various simulation techniques. I gained experimentally validated dynamic atomic structural models of the different states of Ras during the signaling cascade of cell growth. From these atomic models I decoded the catalysis mechanism of GTP hydrolysis by Ras.
My recent research project at the TCBG aims to elucidate structure, dynamics, and function of the 26S proteasome at atomic detail. Therefore, I am improving hybrid structure analysis strategies to combine structural data from sources such as X-ray crystallography and cryo-electron microscopy (cryo-EM) with computational modeling.
Home Department: Beckman Institute for Advanced Science and Technology
Office Address: Beckman Institute, Room 3157
Office Phone: 001-217-244-0177
Email Address: trudack@ks.uiuc.edu
Visit my profile on ResearchGate
Education
- Docctoral Degree in Physics (2013) - Ruhr University of Bochum - Germany
- Diploma Degree in Physics (2007) - Ruhr University of Bochum - Germany
Advisor: Prof. Dr. Klaus Gerwert
Thesis Title: Decoding of the enzymatic catalysis of small GTPases by quantam mechanics and molecular mechanics simulations of time resolved FTIR spectra
Advisor: PD Dr. Jürgen Schlitter
Thesis Title: Development of force field parameter for guanosine triphosphate for molecular dynamics simulations
Research Interests
- Molecular Dynamics
- Cellular Mechanisms
- Protein Degradation
- Signal Transduction
- Hydrolysis Mechanism
- ATPases: Proteasome
- GTPases: Ras
- Integrative Modeling
- Theoretical IR Spectroscopy
Research Highlights

While waste recycling became popular in our daily life more recently, living cells have mastered recycling of their protein content since their very beginning. Recycling of unneeded protein molecules in cells is performed by a molecular machine called 26S proteasome, which cuts these proteins into smaller pieces for reuse as building blocks for new proteins. Proteins that need to be recycled are labeled by tags made of poly-ubiquitin protein chains. The 26S proteasome machine recognizes and binds to these tags, pulls the tagged protein close, then unwinds it, and finally cuts it into pieces. Despite its substantial role in the cells life cycle, the proteasomes atomic structure and function still remain elusive. Employing a combination of computational techniques implemented in NAMD along with cryo-electron microscopy data we obtained an atomic structure of the human 26S proteasome and investigated the mechanism underlying substrate recruitment and unfolding.
More information is provided on our proteasome website
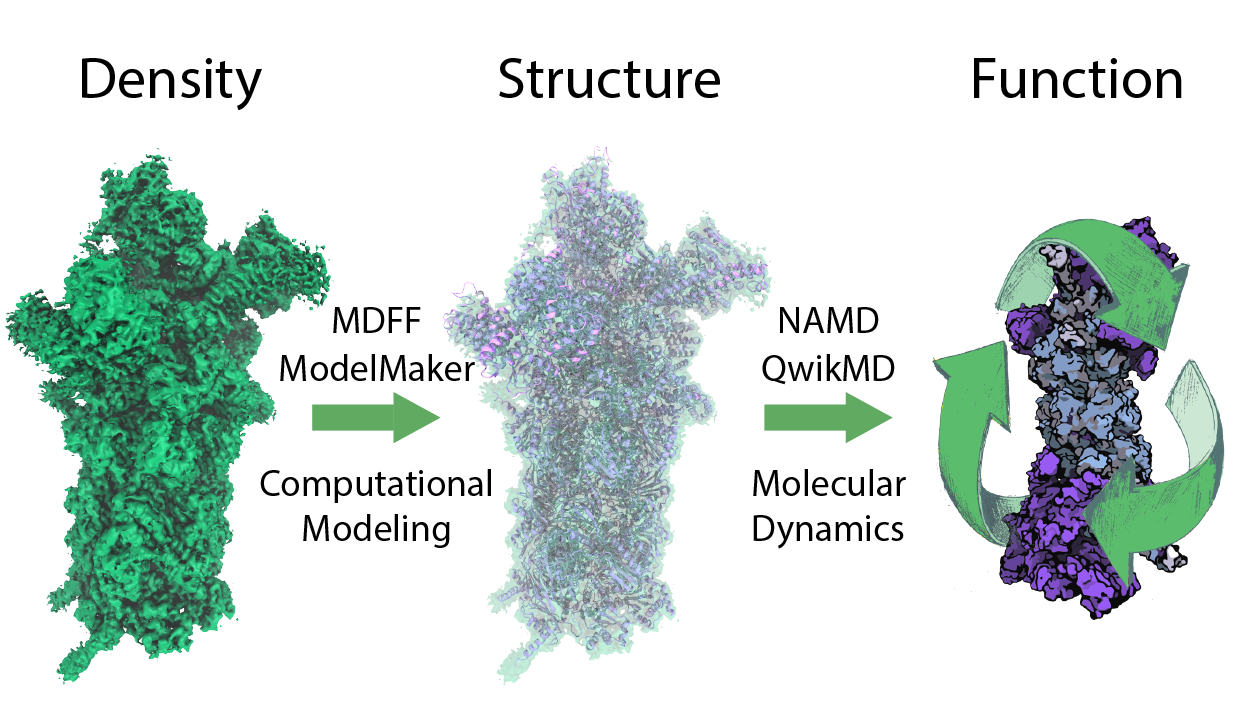
Hybrid structure analysis combines structural data from sources such as X-ray crystallography and cryo-electron microscopy (cryo-EM) with computational modeling and has become a successful strategy for resolving structures of very large macromolecular complexes found in living cells. The computational tools employed aim to automate the process of structure analysis in order to avoid human bias, yet the experience of structural biologists may actually be a desirable factor in structure refinement. Therefore, we developed a hybrid structure analysis strategy that integrates user expertise into an interactive version of model building, the user being guided by experimental data and automated structure prediction. The resulting Rosetta/MDFF tool is of particular value in cases in which structure analysis has to deal with highly flexible or multi-conformational domains. The protocol permits one also to complete missing structural information through de novo structure prediction based on Rosetta.
Everything that living things do can be understood in terms of jigglings and wigglings of atoms.
Richard Feynman's remark in the early 1960's summarizes what is today widely accepted, namely,
that molecular processes can be described by the dynamics of biological molecules, therefore connecting
protein dynamics to biological function. Molecular dynamics (MD) is by far the best tool to investigate
jigglings
and wigglings
of biological systems. Advances in both software and hardware
have spread the use of MD, however the steepness of the learning curve of the methodology of MD
remains high. To assist new users in overcoming the initial barrier to use MD software, and to help the more advanced users to
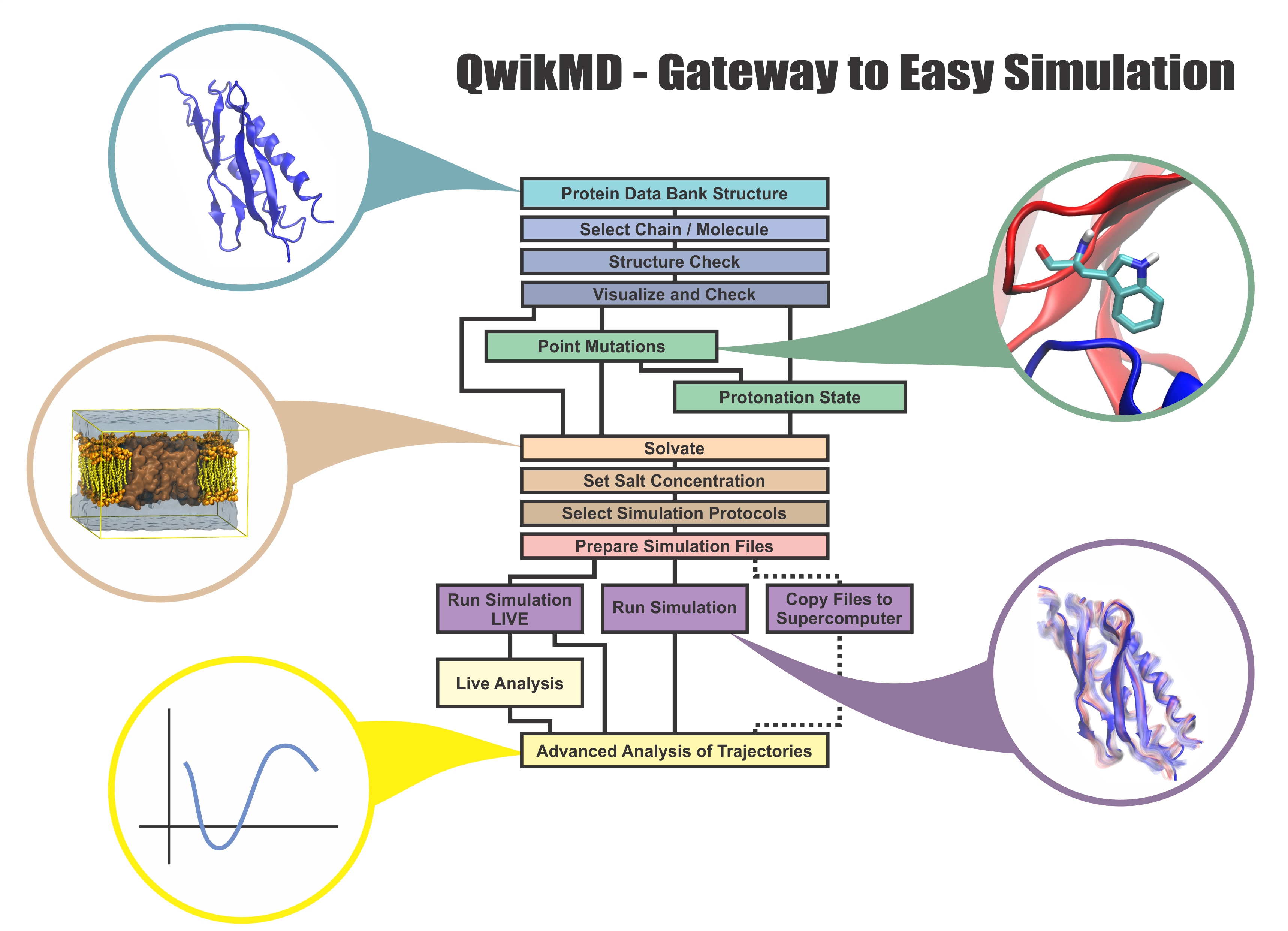
speed up tedious steps, we have developed the QwikMD software, as decribed in a recent paper.
By incorporating an easy-to-use point-and-click user interface that connects the widely used molecular graphics
program VMD
with the powerful MD program NAMD,
QwikMD allows its users to prepare both basic and advanced MD simulations in just a few minutes. At the same time,
QwikMD keeps track of every step performed during the preparation of the simulation, allowing easy reproducibility
and shareability of protocols. More information about QwikMD, as well as introductory tutorials are available on our
QwikMD webpage.
QwikMD is available in VMD 1.9.3 or later versions.
Easy Setup of MD Simulations
Point Mutations
Changes in Protonation
Protocols for MD and SMD
Live View Simulations
Integrated Basic Analysis
Info Buttons to Guide Novices
Advanced Protocols
Membrane Environment
Advanced Analysis
Available on Amazon Cloud
Full log Capabilities
Easy Reproducibility
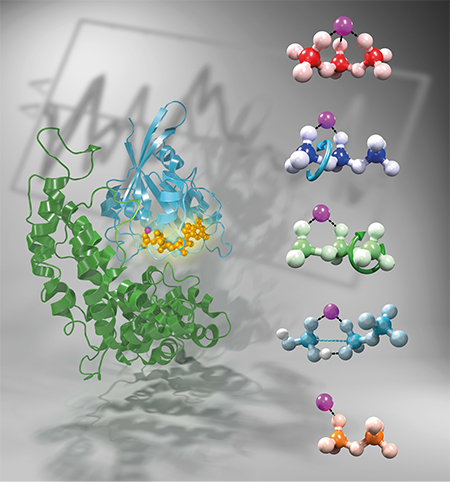
Beside large macromolecular motors I also investigate the enzymatically catalyzed reaction mechanisms of small GTPases. Many cellular processes are regulated by members of the Ras superfamily. They are switched "on" by GDP-to-GTP exchange and "off" by a GTPase reaction. GTP hydrolysis is vitally important for the regulation of several signal-transduction processes in living cells. In the case of Ras, external growth signals are transduced to the nucleus. Site-specific oncogenic mutations inhibit the GTPase reaction and the growth signal can no longer be down-regulated. This contributes to uncontrolled cell growth, eventually leading to cancer. Common to all members of the Ras superfamily is the catalysis of GTP hydrolysis by the G domain. Ras-catalyzed GTP hydrolysis is five orders of magnitude faster than in water (30 min vs. 200 d). However, to control growth signals in the living cell, a further increase of five orders of magnitude in the reaction rate (30 min to 50 ms) is enabled. This is effected by GTPase-activating proteins (GAPs) that interact with Ras. This down-regulation of small GTPases by GAP-catalysis is crucial in the living cell and its interference causes several diseases.
I have developed a workflow to combine the advantages of biomolecular simulations with time-resolved Fourier-transformed infrared (FTIR) spectroscopy measurements and X-Ray crystallographic structural analysis. By this we elucidate the reaction mechanism of the by Ras enzymatically catalyzed GTP hydrolysis and resolve with highest spatio-temporal resolution possible, how this remarkable catalysis is brought about.
My developed workflow combines MM, QM and QM/MM simulations in order to gain detailed information about structure, dynamics and charge distribution of the active site of small GTPases. These computer models are validated by comparing the calculated IR spectra with the experimentally measured ones. This leads to an experimentally validated refinement of the X-ray structure with sub Ãngstroem resolution. Within my work I have proven that sub Ãngstroem resolution of the dynamic structural models is essential to elucidate the catalytic effect of the active site of the small GTPase Ras on GTP hydrolysis. Ras rotates the g-phosphate relative to the b-phosphate from a staggered into an eclipsed conformation and GAP binding further rotates the a- relative to the b-phosphate into an eclipsed conformation. Thereby the oxygen atoms repel each other and GTP is driven in an energetically unfavourable conformation. Furthermore, the protein induced charge shift at the g-phosphate prepares for an optimal nucleophilic attack of the water molecule. Protein binding induce a precatalytic GTP conformation by which the activation free energy is reduced as compared to GTP in water. Beyond, the magnesium ion provides temporary storage for electrons during hydrolysis. All in all, the protein environment drives the conformation and the charge distribution of GTP closer to that of its hydrolysis transition state and facilitates thereby hydrolysis. This result exemplified, that understanding protein catalysis deserves time-resolved studies at atomic details provided here by combining vibrational spectroscopy and biomolecular simulations.
Furthermore, I have been able to resolve the detailed structure and inclusion into the protein environment of the nucleotide during a transient intermediate state within a cooperation project with the Chines Academy of Science Max Planck Gesellschaft Partner Institue for Computational Biology (CAS MPG PICB) in Shanghai (China). In addition, I have been able to show that the clustering of Ras at a membrane is driven by dimerization. This was the result of a combination of MD simulations, attenuated total reflection fourier transform infrared (ATR FTIR) spectroscopic investigations and Foerster Resonance Energy Transfer (FRET) experiments.
Collaborators
- Max Planck Institute of Biochemistry - Department of Wolgang Baumeister - Germany
- Max Planck Institute for Chemical Energy Conversion - Department of Frank Neese - Germany
- Quantum Biology and Computational Physics Group - University of Southern Denmark - Denmark
- Biochemistry Center - University of Heidelberg - Germany
- Department of Biophysics - Ruhr University Bochum - Germany


Publications
- Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Wehmer M.*, Rudack T.*, Beck F.*, Aufderheide A, Pfeifer G., Plitzko J.,Foerster F., Schulten K., Baumeister W., Sakata E.
- Local Mode Analysis: Decoding IR spectra by visualizing molecular details. Massarczyk M.*, Rudack T.*, Schlitter J., Kuhne J., Koetting C., Gerwert K.
- The structure of the 26S proteasome at a resolution of 3.9 A. Schweitzer A.*, Aufderheide A.*, Rudack T.*, Beck F., Pfeifer G., Plitzko J., Sakata E., Schulten K., Foerster F., Baumeister W.
- QwikMD Integrative Molecular Dynamics Toolkit for Novices and Experts. Ribeiro J. V.*, Bernardi R. C.*, Rudack T.*, Stone J. E., Phillips J. C., Freddolino P. L., Schulten K.
- Recognition of poly-ubiquitins by the proteasome through protein re-folding guided by electrostatic and hydrophobic interactions. Zhang Y.*, Vukovic L.*, Rudack T.*, Han W., and Schulten K.
- Computational methodologies for real-space structural refinement of large macromolecular complexes. Goh B. C., Hadden J. A., Bernardi R. C., Singharoy A., McGreevy R., Rudack T., Cassidy C. K., and Schulten K.
- Catalysis of GTP hydrolysis by small GTPases at atomic detail by integration of X-ray crystallography, experimental and theoretical IR spectroscopy. Rudack T.*, Jenrich S.*, Brucker S., Vetter I. R., Gerwert K., Kötting C.
- Molecular dynamics simulations of large macromolecular complexes. Perilla J. R., Gob B. C., Cassidy C. K., Liu B., Bernardi R. C., Rudack T., Yu H., Wu Z, Schulten K.
- Die Geheimnisse des Lebens berechnen. Rudack T., Perilla J. R., Schulten K.
- Ras and GTPase-activating protein (GAP) drive GTP into a precatalytic state as revealed by combining FTIR and biomolecular simulations. Rudack T., Xia F., Schlitter J, Kötting C., Gerwert K.
- Detailed Structure of the H2PO4 Guanosine Diphosphate Intermediate in RasGAP Decoded from FTIR Experiments by Biomolecular Simulations. Xia F.*, Rudack T.*, Cui Q., Kötting C., Gerwert K.
- The Role of Magnesium for Geometry and Charge in GTP Hydrolysis, Revealed by Quantum Mechanics/Molecular Mechanics Simulations. Rudack T., Xia F., Schlitter J., Kötting C., Gerwert K.
- N-Ras forms dimers at POPC membranes. Güldenhaupt J.*, Rudack, T.*, Bachler P., Mann D., Triola G., Waldmann H., Kötting C., Gerwert K.
- Exploring the Multidimensional Free Energy Surface of Phosphoester Hydrolysis with Constrained QM/MM Dynamics. Li W., Rudack T., Gerwert K., Gräter F., Schlitter J.
- The specific vibrational modes of GTP in solution and bound to Ras: a detailed theoretical analysis by QM/MM simulations. Xia F., Rudack T., Kötting C., Schlitter J., Gerwert K.
Visit my Google Scholar profile
PNAS 2017 114(4):1305-1310.
The Journal Physical of Chemistry B, Klaus Schulten Memorial Issue 2017 121(15):3483-3492.
PNAS 2016 113(28):7816-7821.
Nature Scientific Reports 2016 6:26536
The Journal of Physical Chemistry B, McCammon Festschrift 2016 120(33):8137-8146.
Annual Review Biophysics, 2016 in press.
Journal of Biological Chemistry 2015 290 (Cover)
Current Opinion in Structural Biology 2015 31:64-74.
Spektrum der Wissenschaften 2014 11:86-95.
Proceedings of the National Academy of Science of the USA 2012 109(38):15295-15300.
Journal of the American Chemical Society 2012 136:20041-20044.
Biophysical Journal 2012 103:293-302.
Biophysical Journal 2012 103:1585-1593 (Cover).
Journal of Chemical Theory and Computation 2012 8 (10):3596-3604.
Physical Chemistry Chemical Physics 2011 13:21451-21460.
* equal contribution
Awards
- 2016 Attendance Fellowship of the German Accademic International Network (GAIN) to the 16th Annual GAIN Meeting 2016, Washington, D.C., US
- 2015 Travel Award of the German Biophysical Society to the European Biophysical Society Meeting 2015, Dreseden, Germany
- 2015 Best Image Award of the German Biophysical Society
- 2014 Feodor-Lynen Postdoctoral Fellowship of the Alexander von Humboldt Foundation
- 2014 Nominated for the best interdisciplinary doctoral thesis of the Ruhr-University Bochum
- 2012 Cold Spring Habor Asia Fellowship: Best Poster Award
Ras and GAP drive the geometry and charge of GTP into a precatalytic state as revealed by combining FTIR and QM/MM simulations,
2012 Cold Spring Harbor Asia Conference: Small GTPases at Different Scales: Proteins, Membranes, Cells, Suzhou, China. - 2011 Best Poster Award of the European Biophysical Society Mg2+ is a temporary electron storage during the GTP Hydrolysis Mechanism, 8th EBSA European Biophysics Congress, Budapest, Hungary.
- 2009 Travel Award of the German Biophysical Society to the European Biophysical Society Meeting 2009, Genua, Italy\\
Talks
- 03/25/2017 ModelMaker: A Tool for Interactive Modeling of Complete Proteins Guided by CryoâEM,
Structure Prediction, and Molecular Dynamics.
Computer Simulation and Theory of Macromolecules 2017. Huenfeld, Germany - 03/08/2017 The story of a "precise" proteasome model: Integrating experimental results and user expertise into computational modeling.
Ringberg Meeting of the Department of Molecular Structural Biology MPI of Biochemistry, Ringberg, Germany - 02/25/2017 From atom to cell: Integrating experimental results and user expertise into computational modeling.
Structural Transitions of Biomolecules in Experiment and Theory, Joint meeting of the Czech and German Biophysics Societies, Huenfeld, Germany - 02/25/2017 From atom to cell: Integrating experimental results and user expertise into computational modeling.
Structural Transitions of Biomolecules in Experiment and Theory, Joint meeting of the Czech and German Biophysics Societies, Huenfeld, Germany - 07/14/2016, How is chemical energy converted into motor action? Integrating experimental methods into computational modeling can provide answers.
Max Planck Institute for Chemical Energy Conversion, Department of Frank Neese, Germany - 07/12/2016, Integrating experimental results and computational modeling unravels functional details about catalytic centers of large macromolecular complexes.
Collaborative Research Center 642 seminar, SFB TR83, SFB TR186 Guest Seminar, Biochemistry Center of the University Heidelberg (BZH), Germany - 06/15/2016, Molecular Dynamics Simulations of Large Macromolecular Complexes.
NCSA Blue Waters Symposium for Petascale Science and Beyond, Sunriver, US - 05/06/2016, Molecular Dynamics Simulations for Everyone From iPad to Supercomputers From Atom to Cell.
Collaborative Research Center (SFB) 642 seminar, Ruhr-University Bochum, Germany - 05/04/2016, How the Ras protein became a wind-up car.
Anniversary Symposium for Klaus Gerwert, Ruhr-University Bochum, Germany - 03/04/2016, Integrating experimental results and user expertise into computational model building.
Amaro group, UCSD, San Diego, US - 03/03/2016, Integrating experimental results and user expertise into computational model building.
Lander group, Scripps Research Institute, San Diego, US - 03/01/2016, A tool to integrate user expertise into building atomic level models for large bimolecular systems.
60th Biophysical Society meeting, LA convention Center, US - 09/10/2015, Exploring GTPase catalysis at atomic structure level by combining biomolecular simulations with FTIR spectroscopy.
2015 European Conference on the Spectroscopy of Biological Molecules (ECSMB), Bochum, Germany - 02/09/2015, Integrating experimental results and user expertise into computational model building.
Pande Group, U Stanford, San Francisco, US - 01/09/2015, Integrating experimental results and user expertise into computational model building.
Martin Group, UC Berkeley, San Francisco, US - 08/31/2015, Integrating experimental results and user expertise into computational model building.
Sali Group, UCSF, San Francisco, US - 07/27/2015, Rosetta/MDFF: A tool to integrate userÕs expertise into cryo-EM model building applied to the 26S proteasome.
Baumeister group, Max Plank Institute for Biochemistry, Munich, Germany - 07/20/2015, Ubiquitin recruitment and transport through the 26S proteasome unraveled by an orchestra of MD simulation algorithms.
2015 European Biophysical Society Meeting, Dresden, Germany - 09/28/2012, Decoding structural information from FTIR spectra by biomolecular simulations.
Cold Spring Harbor Asia Conference: Small GTPases at Different Scales: Proteins, Membranes, Cells, Suzhou, China - 05/18/2012, Ras and GAP drive GTP into a precatalytic state: a combined FTIR and QM/MM simulation study.
CAS-MPG Partner Institute for Compuational Biology (PICB), Shanghai, China - 04/20/2012, Catalysis of GTP Hydrolysis by 10 Orders of magnitude by Ras-Ras$\cdot$GAP Revealed at Atomic Detail by Combining QM/MM Simulations and FTIR Spectroscopy.
Computer Simulation and Theory of Macromolecules 2012, Hunfeld, Germany - 09/08/2011, Mg2+ is a temporary electron storage at GTP Hydrolysis.
2nd summer school of the graduate college of the SFB 642, Wenden, Germany - 05/20/2011, The Ras Protein: Theoretical IR Spectroscopy of Different States of GTP Hydrolysis.
Spectroscopy Symposium, CAS-MPG Partner Institute for Compuational Biology (PICB), Shanghai, China - 08/16/2010, Biomolecular Simulations on the Ras Protein.
Symposium on Biomolecular Simulations, CAS-MPG Partner Institute for Compuational Biology (PICB), Shanghai, China - 04/16/2010 Hydrolysis Mechanism of Ras Investigated by QM/MM Simulations, Computer Simulation and Theory of Macromolecules 2010. Hunfeld, Germany
- 01/25/2010, QM/MM Simulations on the Hydrolysis Mechanism of Ras.
B-IT/PICB Workshop: Computational Life Science, Bonner-IT, Bonn, Germany - 05/23/2009 Interaction of N-Ras-GTP and N-Ras-GDP with a POPC Bilayer.
Protein-Protein-Interaction: Experiment and Theory, Joint meeting of the Danish and German Biophysics Societies, Hunfeld, Germany - 03/02/2009 Interaction of N-Ras-GTP and N-Ras-GDP with a POPC Bilayer.
Bioquant, University Heidelberg, Heidelberg, Germany



