Molecular Basis for Anthrax Intoxication
Anthrax is caused by Bacillus anthracis, a spore-forming bacterium that can survive extreme conditions over long period. Once the bacterium enters an animal or a human host, the spores germinate and reproduce bacteria disseminated rapidly to the blood stream. In order to suppress the immune responses of the host and promote the survival of the bacterium, Bacillus anthracis secretes anthrax toxin composed of three types of killer proteins: protective antigen (PA), lethal factor (LF), and edema factor (EF). The toxin triplet targets primarily macrophages, defensive cells that ingest invading microorganisms.
The assault to a target cell by anthrax toxin is an "organized" crime. Individually, none of the three proteins is toxic. In order to function, LF or EF must team up with PA, which is responsible for transporting LF and EF into a target cell. Once inside the cell, LF converts ATP to cyclic AMP, while EF disables MAPKKs, a family of signaling proteins. These attacks disrupt various cellular signaling pathways of macrophages and some other cells, essentially shutting down the host's immune system and often leading to death of the host.
Killers' Entry Route
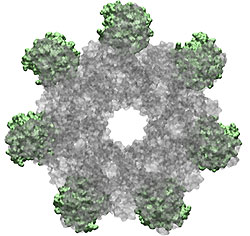
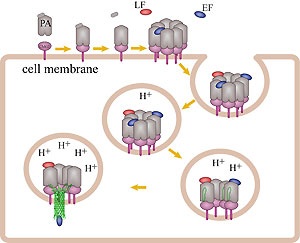
How do the anthrax toxin killers penetrate a membrane wall, the defense line of a host cell? It turns out that the toxin takes an intricate entry route: As the first step, PA, the component of toxin responsible for transporting poisonous LF and EF, hijacks a receptor protein on the cellular surface. A small piece of PA is subsequently cut off by a protease, an enzyme used by the host to degrade proteins. The remaining receptor-bound portion of PA assembles into a ring-like structure with six other PA-receptor pairs. The cleavage exposes anchor sites for LF and EF, attaching to the top side of PA away from the receptor. The killers somehow delude the cell, which mistakenly uptakes the entire toxin-receptor complex through a process known as endocytosis. The resultant structure, an endosome, is a bubble-like compartment enclosed by membrane with the toxin-receptor complex sitting inside. While endosome diffuses and moves close to the targets of LF and/or EF, the complex undergoes conformational changes and forms a barrel-like pore inserted into the membrane of the endosome. Through the pore, the toxic LF and EF are translocated and released to attack their targets within the cell.
Toxin Pore Formation
Although the general picture of the anthrax intoxication pathway has been established, many details still remain a mystery. One key question is how the heptameric PA-receptor complex initiates formation of the pore for ferrying the toxic cargo. Structurally, each PA has four domains: I-IV. It is believed that each of the seven PA domain IIs contributes one long loop lining the eventual 14-beta-stranded barrel-like pore. However, according to recent crystal structures of PA-receptor complexes resolved in a stage prior to pore formation, the movement of this PA loop is constrained because PA domain II (PAII), together with domain IV (PAIV), binds to the receptor, capillary morphogenesis protein 2 (CMG2). In order to liberate the loop for the pore formation purpose, PAII must separate from the complex, further dislodge a few loops and unfold a couple of beta-strands as well. On the other hand, it has been known that low acidic conditions are required for pore formation. Studies have found that the conversion from a pre-pore structure to a pore-containing structure proceeds under acidic condition between pH 5 and 6. Therefore, it is attempting to hypothesize that the acidified environment of endosome triggers the dissociation of PAII for pore formation.
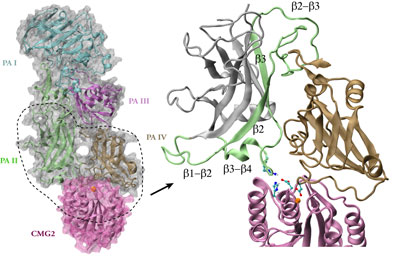
Structure of anthrax toxin PA in complex with the headpiece of a host cellular receptor CMG2. Under acidic conditions, PA unravels part of its domain II, releasing peptides for pore formation.
The hypothesis can be examined with molecular dynamics simulations that provide important structural insights into the pore-formation mechanism. Simulations revealed that under neutral pH conditions PAII orients one loop so that a positively charge amino acid (Arg344PA) interacts optimally with a negatively charged amino acid (Glu122CMG2) on the receptor. The interaction between the pair, a so-called salt bridge, prevents the toxin from dissociation. Acidic conditions induce the addition of a proton to certain types of protein amino acids like glutamates and histidines, a chemical reaction known as protonation. In many cases, protonation of few amino acids at the interface between a ligand-receptor pair severely disrupts their interactions because of the introduction of additional charges. Indeed, atomic simulations demonstrated that the protonation of two amino acids involving or neighboring the salt bridge (His121CMG2 and Glu122CMG2) triggers rapid dissociation of PA domain II from the receptor. The trio, His121 and Glu122 of CMG2, and Arg344 of PA, appear to serve as a pH-sensitive switch that controls the pore formation.
- Click here for a movie (mpeg, 3.5M) showing a crucial salt bridge stabilized at the binding interface between the anthrax toxin PA (green) and its receptor CMG2 (pink).
- Click here for a movie (mpeg, 3.6M) showing the dissociation of PA domain II upon the protonation of His121 and Glu122 of CMG2.
Toxin Receptor Recognition
How does anthrax toxin PA recognize its receptors? Before the crystal structures of a PA-receptor complex became available, sequence analysis suggested that PA receptors contain a structural motif known as the metal ion dependent adhesion site, or MIDAS. At this site, a receptor acquires a divalent cation, typically Mg2+, which grips a positively charged amino acid from its ligand. The MIDAS motif has been identified in other cellular receptors best characterized by integrins, a family of proteins relaying chemical and mechanical signals across membrane through conformational changes. How does PA recognize specifically its receptor like CMG2 but not integrins? The puzzle was solved in the crystal structures of the PA-CMG2 complex, where a second binding site between PA domain II and CMG2 was unveiled, along with the MIDAS interactions identified between PA domain IV and the receptor. While the configuration of the MIDAS motif on CMG2 exhibits the same "open" (ligand-bound) form as a typical integrin, the second binding site for PAII is conserved only between anthrax receptors, but does not exist in integrins. This differentiates cellular receptors targeted by anthrax toxin from integrins.
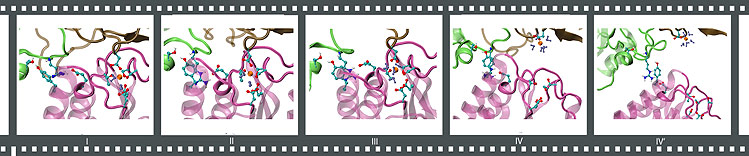
Forced separation of an anthrax toxin-receptor complex under neutral conditions.
- Click here for a movie (mpeg, 4.2M) showing stretching the receptor CMG2 away from the anthrax toxin protein PA under neutral conditions.
- Click here for a movie (mpeg, 5.0M) showing stretching the receptor CMG2 away from the anthrax toxin protein PA after PA domain II becomes partially dissociated under acidic conditions.
By means of steered molecular dynamics, key structural components that are crucial for maintaining the stability of a ligand-receptor pair can be identified. The anthrax toxin-receptor complex PA-CMG2 has a binding affinity hundreds of times higher than a typical integrin-ligand complex. A clue for how the anthrax toxin-receptor complex achieves a high affinity state has emerged by comparing the following two scenarios: (a) stretching a neutral structure vs. (b) stretching an acidic structure where PA domain II becomes dissociated (check out the movie clips above). In both cases, simulations of stretching the PA-CMG2 complex have shown that the MIDAS interactions play a major role in stabilizing the binding because the resultant peak force consistently correlates with the disruption of these interactions. However, in scenario a, the peak force is significantly lower than scenario b due to the absence of PAII-CMG2 interactions. A short loop links the MIDAS site to the PAII binding site in the receptor. Upon the dissociation of PAII under acidic conditions, the loop become more flexible, cascading conformational changes that disturb the coordination to the MIDAS cation. As a result, disrupting the MIDAS interactions in the acidic structure requires a much lower peak force than that for disrupting the intact MIDAS interactions exhibited in the neutral structure. This observation corroborates the hypothesis that the binding between PA domain II and the receptor dramatically increases the stability of the complex, and provides an explanation for how the complex achieves a high affinity state through the tight coordination of the MIDAS cation.
Publications
Investigators
Related TCB Group Projects
Page created and maintained by Mu Gao.