MDX
Replacement of MDX by NAMD-Lite
The name "MDX" is being replaced by "NAMD-Lite" to better denote the role of this project as a vehicle for NAMD method development. The new version of this software is available from the NAMD-Lite webpage.
Recent News
- Added support in MDX for reading CHARMM parameter files, plus some bug fixes, released as version 1.0.2.
- The MDX Tutorial is now complete.
- The "NAMD-lite" announcement has been emailed to the CCL.
- Added a second demo to MDX, released as version 1.0.1.
- "NAMD-lite" Announcement
- MDX Slide Presentation (PDF)
Overview
MDX is a collection of C libraries to enable the development of methods for molecular dynamics of biomolecules. The target audience is anyone (computer scientist or physicist) interested in using and extending these libraries for the purpose of methods development and experimentation.
MDX provides a modular approach to developing molecular dynamics software. The goal is to develop reusable modules with well-defined interfaces towards the implementation of a sequential molecular dynamics program and related tools. The priority is to create MD codes that are easy to understand and modify, enabling straightforward design and testing of new methods. Flexible software libraries will provide tools for taking care of common tasks, allowing more rapid development of revolutionary techniques.
Two major methods currently being developed are multilevel summation method (MSM) for the fast evaluation of electrostatic forces and MDAPI, a high-performance interfacing of a front end with an MD engine, intended for use in a future release of NAMD.
The codes have been successfully tested and should compile and run on Linux, Solaris, and Darwin (Mac OS X). Compiling the software requires the GNU version of make.
Some Details
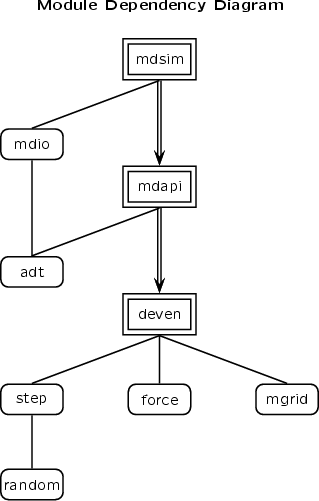
MDSim Program
This executable program from MDX performs an MD simulation using a subset of NAMD configuration options, so can be viewed as "NAMD-lite". The mdsim (front end) uses the MDAPI to interface to deven (MD engine), providing a non-trivial demonstration of the interface.
MDSim performs constant energy simulation using leapfrog (velocity Verlet) time integration. It can simulate nonperiodic or fully periodic orthogonal cells using a cutoff for electrostatics. It can also make use of mgrid (MSM library) to compute long-range electrostatics, but this feature is restricted to cubic cells.
Libraries
- force - Evaluates bonded potentials, cutoff nonbonded potentials, nonbonded exclusions, and certain boundary constraints. Supports orthogonal cells that may be periodic in 0, 1, 2, or 3 dimensions. The different force types can be enabled individually or collectively.
- mgrid - MSM restricted to cubic cells that are either nonperiodic or fully periodic.
- step - Performs leapfrog (velocity Verlet) time integration. Can also initialize velocities from a desired temperature distribution and calculate kinetic energy and temperature reductions.
- random - Generates random numbers. Used by the step library.
- mdapi - Interfaces a front end with an MD engine. This version is single-threaded, considered to be the reference implementation of the MDAPI specification.
- mdio - Molecular dynamics I/O classes. Reads NAMD-style configuration files, X-Plor and CHARMM parameter files, and PSF molecular structure files. Reads and writes coordinate PDB files and NAMD-style binary restart files. Writes DCD files (less than 2 GB).
- adt - Container classes used by MDIO and MDAPI.
- deven - MD engine that utilizes force, mgrid, and step libraries.
- simen - Simple MD engine that utilizes force library.
Caveats
For a scientist interested in using these codes for methods development, it is not necessary, and probably not desirable, to comprehend the MDAPI. The other major libraries (step, force, mgrid, and mdio) are much more accessible and can lend themselves more quickly to methods development, plus they contain all of the starting pieces needed for MD codes. One thing to note is that all of these libraries depend on the molecular and force field data types defined in the src/mdapi/mdtypes.h header file, which is well-commented. Reading Chapter 2 of the MDAPI Specification (doc/mdapi.pdf), in which these data types are discussed, might also be helpful.
This code base is under active development. Even a more mature module, such as the MDAPI, is subject to experience some changes to its interface as some design issues are resolved.
Future Plans
List of planned MSM library enhancements:
- Optimization for "sparse" nonperiodic domains to cut away corners from cube where atoms do not exist.
- Extend nonperiodic systems to rectangles.
- Permit grid sizes for periodic domains to be different sized powers of 2 between dimensions or non-powers of 2 (e.g. allow a factor of 3 or 5).
- Add support for semi-periodic cells.
- Extend periodicity to rectangles.
- Extend periodicity to parallelepipeds.
- Perform fast reinitialization of constants for domain dimension updates (needed for flexible cells for constant pressure simulation).
Other plans to extend MDX include:
- Incorporation of Wei Wang's polarizability code.
- Implementation of multiple time stepping.
- Computation of "shadow" Hamiltonians.
Updates to the documentation:
- MDAPI needs to be "doxygen-ated", particularly to move the reference section of the MDAPI Specification so that it is maintained directly within the header files.
Contact
If you have questions please send email to the author David J. Hardy at dhardy@ks.uiuc.edu.
Source Code and Documentation
The documentation for the libraries and the specification for the MDAPI are included within this source code distribution.
- Download MDX Version 1.0.2 (source code distribution)
There is also online web documentation available for the main libraries and the MDAPI Specification document.
- MDX Library Web Documentation (online HTML)
- MDAPI Specification, Version 0.9 (PDF)
Changes in version 1.0.2
- Supports reading CHARMM parameter files. Agrees with NAMD on bpti. (There is no need to use the NAMD "paraTypeCharmm" directive, as these files are recognized automatically based on the leading '*' character for the header comments.)
- Correctly implements NAMD's support for X-Plor parameter files with dihedral multiplicities indicated by the X-Plor PSF file. Agrees with NAMD on apoa1.
- Fixed bug correctly matching dihedrals with multiplicities.
- Fixed memory allocation in array.c when initializing length zero array. Credit to Alessandro Pedretti for this bug fix.
Changes in version 1.0.1
- Includes a second demonstration that illustrates use of multilevel summation method for computing electrostatics.
License
University of Illinois/NCSA Open Source License
Page created and maintained by David J. Hardy.



