RNA Exosome
Introduction
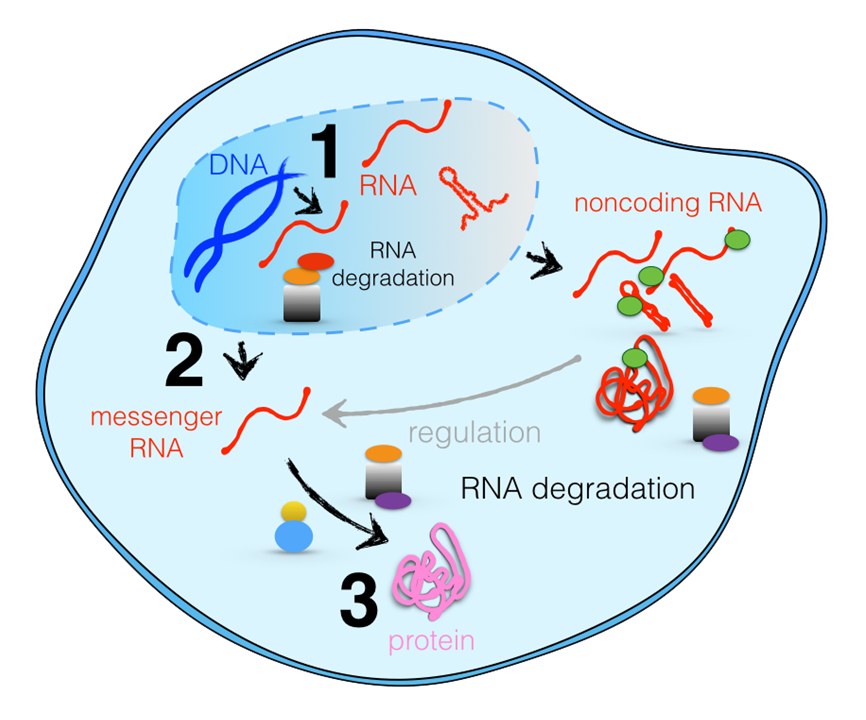
Function and development of every organism is encoded in DNA of its genome. A large fraction of DNA is copied into RNA molecules that can execute the instructions stored in DNA. The operation of the cell has to be tightly controlled, so that synthesized molecules contain no errors and exist for appropriate amounts of time at appropriate concentrations. To achieve a tight regulation of the RNA content in the cell, proteins and protein complexes act as molecular machines and recognize and degrade RNA transcripts whenever needed. The RNA exosome complex is a major RNA degradation complex in eukaryotes, responsible for most of 3' to 5' RNA degradation, and is involved in the degradation of multiple RNA types.
The exosome complex can recognize and degrade RNA substrates in vitro even without external energy input. A key and only subunit of the exosome that has the ability to processively degrade RNA, both within the exosome and by itself, is the protein subunit Rrp44, a 3' exonuclease. Recent crystal structures and cryo-electron microscopy maps have revealed the architecture of the exosome complex and its binding mode to RNA during the degradation process. Three main regions, including the cap, the core, and the Rrp44 subunit, assemble into the exosome complex, while forming a long internal channel within it. ssRNA is then channeled within the exosome from cap proteins at the channel entry to the Rrp44 exonuclease active site at the channel exit.
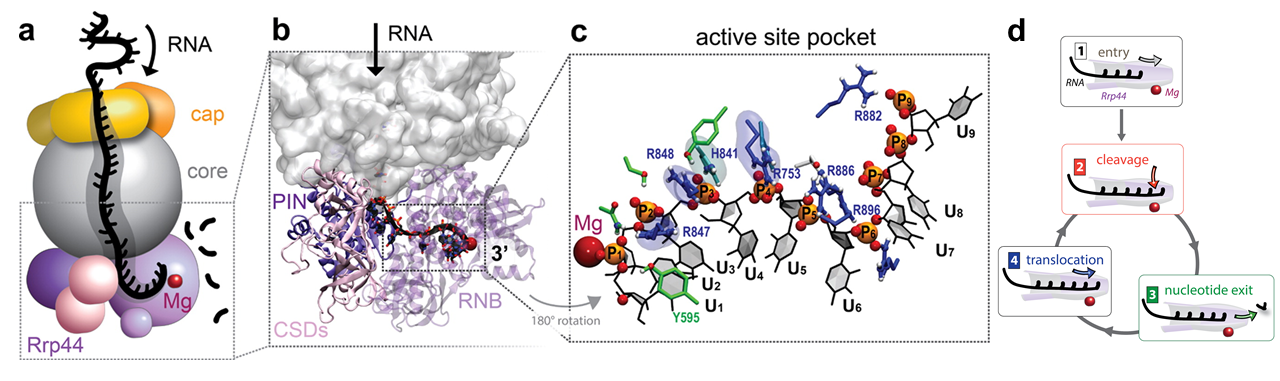
The cap and core subunits of the exosome initially recruit RNA substrates that have ssRNA extensions that can reach Rrp44. Then, the Rrp44 subunit can act both as a motor, by unwinding and pulling RNA substrates toward the cleavage site at the end of the interior channel, and as an enzyme, by hydrolytically cleaving RNA substrates into single nucleotides. Upon entry of RNA into the active site of Rrp44 (step 1), the following steps of the RNA degradation cycle, which involve enzyme and motor actions of Rrp44, are shown schematically in Figure 3: (step 2) The 3'-end RNA nucleotide is hydrolytically cleaved at the Rrp44 exonuclease active site; (step 3) the cleaved nucleotide leaves the active site; and (step 4) RNA translocates forward by one step, moving its 3'-end again into the position required for cleavage. The final products of RNA degradation by Rrp44 are 3-5-nt RNA pieces, which are released into the solution; the 4-nt final product is the most abundant.
Processive RNA Translocation in the Exosome
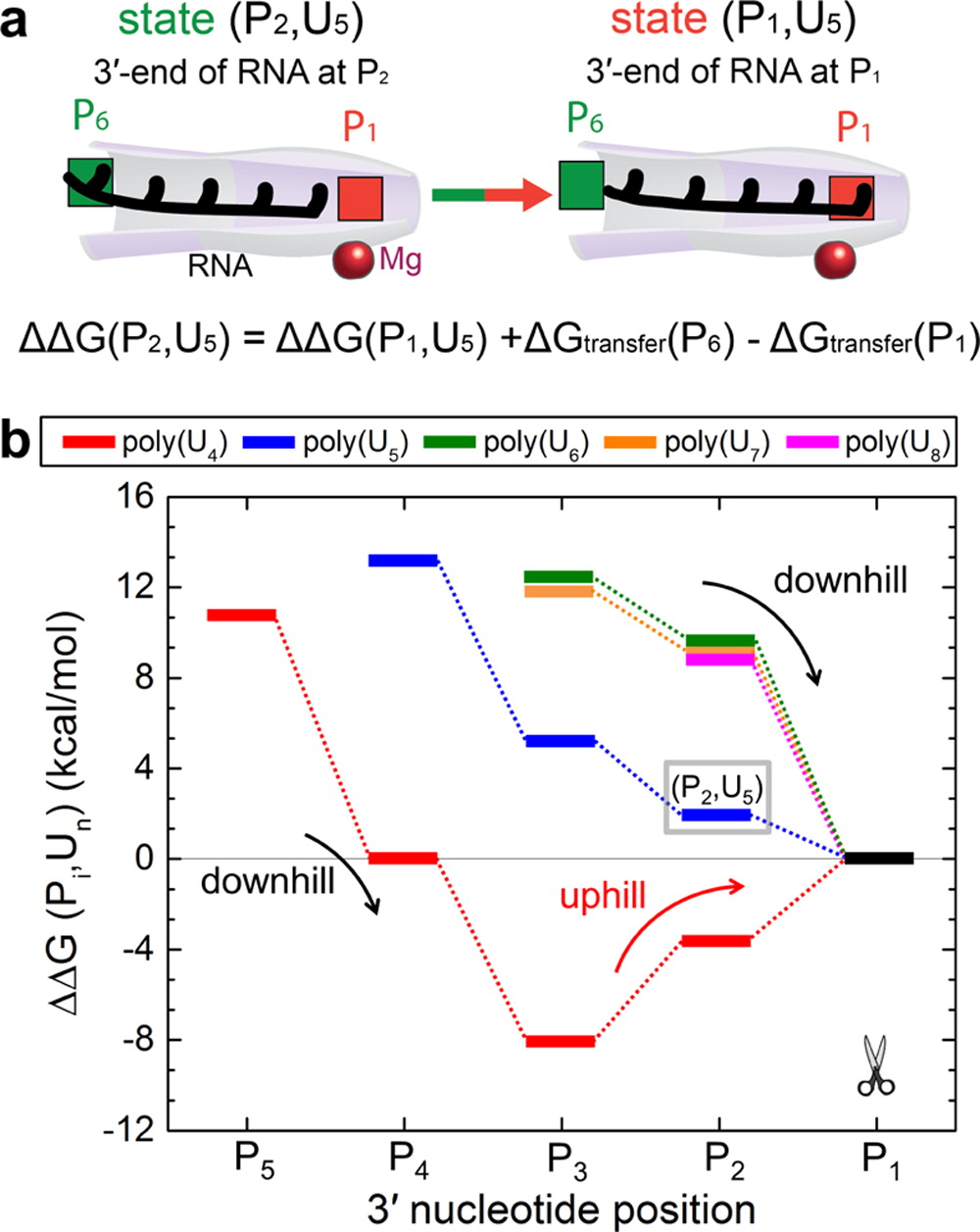
The experimentally determined structure was examined in detail within molecular dynamics simulations. To investigate the factors that contribute to processive translocation and cleavage of RNA in Rrp44 and the exosome, the simulations determined the strength of interactions between RNA and the Rrp44 in the form of transfer free energies of single nucleotides from water to selected positions within Rrp44. Transfer of nucleotides from water to the protein environment is favorable within the whole active site tunnel of Rrp44; the transfer free energies range from -5 to -19 kcal/mol. The exonuclease active site tunnel contains several sites binding RNA very strongly, where the transfer free energies are as large as -14 kcal/mol, akin to the energy gained through ATP hydrolysis.
Exosome function involves continuous translocation of the whole RNA toward the cleavage site. Therefore, the free energy as a function of RNA translocation in the active site tunnel of Rrp44 was determined. The calculations determined that initial entries of poly(U) RNAs are energetically favorable. Furthermore, 5-8-nt RNA segments always have last translocation steps that are accompanied by downhill free energy profiles (step 4 of the cleavage cycle). These steps are thus also energetically favorable. However, the extent of the free energy gain in the last translocation step is length dependent. Interestingly, the free energy profile for translocation of the shortest piece of RNA examined, U4, exhibits a different trend. A downhill free energy profile is observed for U4 entering the active site tunnel to the position where its 3'-end is at position P3. After that point, the profile changes slope, and an uphill free energy profile is observed for translocation of U4 toward the position of cleavage, estimated at 8 kcal/mol. This uphill free energy profile indicates that translocation of U4 RNA into position for cleavage is not energetically favorable.
The simulations identified that two strong RNA binding sites contribute to the active site tunnel of Rrp44 having a strong grip on RNA. These sites also lead to discrimination in length of RNA substrates. The strong RNA binding sites are spatially separated, at positions P1 (the catalytic center) and at positions P4 and P5. RNA substrates that are 4-nt long cannot span both strong binding sites and therefore cannot simultaneously bind to both sites. In fact, simulations show that 4-nt long RNA thermodynamically favors binding to P4 and P5, leading to release of the 4-nt final RNA product, as observed in experiments.
TCBG Publications
Molecular mechanism of processive 3' to 5' RNA translocation in the active subunit of the RNA exosome complex. Lela Vukovic, Christoph Chipot, Debora Makino, Elena Conti and Klaus Schulten. J. Am. Chem. Soc. , 138:4069-4078, 2016.



