TCBG Software at SC2003
NIH National Center for Research Resources Booth Presentations
Title:
NAMD, VMD, and BioCoRE
Location:
NIH NCRR booth
Full Information:
http://nbcr.ucsd.edu/SC2003_NCRR.htm
Times:
3pm Wednesday, 2pm Thursday
Investigators:
Klaus Schulten, Robert Skeel, Laxmikant Kale
Developers:
James Phillips, John Stone, Robert Brunner, Kirby Vandivort, Michael Bach, Justin Gullingsrud, Gengbin Zheng, Paul Grayson
Presenters:
James Phillips and Robert Brunner
Contacts:
Peter Arzberger at UCSD
TCBG Support:
Prepare and present demo.
What will be demonstrated:
We will demonstrate NAMD, VMD, and BioCoRE. NAMD, recipient of a 2002
Gordon Bell Award, is a parallel molecular dynamics code for large
biomolecular systems. VMD is a molecular visualization program for
displaying, animating, and analyzing large biomolecular systems using
3-D graphics. BioCoRE is a collaborative work environment for
biomedical research.
NAMD, VMD, and BioCoRE are developed by the Theoretical and Computational Biophysics Group at the
University of Illinois. NAMD, VMD, and BioCoRE development is supported by
the NIH National Center for Research Resources
and the National Science Foundation.
BioCoRE Presentation in NCSA Booth
Title:
BioCoRE: A Collaborative Work Environment for Biomedical Research, Research Management and Training.
Location:
NCSA booth
Times:
12:30pm Thursday
Investigators:
Klaus Schulten, Robert Skeel, Laxmikant Kale
Developers:
Robert Brunner, Kirby Vandivort, Michael Bach
Presenters:
Robert Brunner
Contacts:
Bill Bell at NCSA
TCBG Support:
Prepare and present demo.
What will be demonstrated:
A resource-centered platform, BioCoRE offers scientists, working together or alone, a seamless interface to a broad range of local and remote technologies such as discipline-specific and general tools, data, and visualization solutions. BioCoRE provides a collection of web-based tools which simplify the tasks that make up the research process, among them co-authoring papers and other documents, running applications on supercomputers, sharing molecular visualization over the Internet, notifying project team members of recent project changes by email, chatting, keeping a lab book, and other practical features. Our presentation will prominently feature the BioCoRE Job Management system, which allows researchers to run computational jobs on various supercomputer systems as well as their own computational resources, using an easy web-based interface. We will also demonstrate how tools like the BioCoRE Control Panel, the Message Board, and the Website Library allow researchers working together but separated geographically to work more efficiently. We willĘ show how the shared file system component of BioCoRE, the BioFS, simplifies the sharing of data and publications, and how BioCoRE can work with the molecular visualization program VMD to allow researchers to share graphical displays of molecular models.
Sun Microsystems Sun Fire Visual Grid Demo
Title:
VMD: Visual Molecular Dynamics
Location:
Sun Microsystems booth
Times:
various, unknown
Investigators:
Klaus Schulten, Robert Skeel, Laxmikant Kale
Developers:
John Stone, Justin Gullingsrud, Gengbin Zheng, Paul Grayson
Presenter:
Sun personnel
Contacts:
Travis Bryson at Sun Microsystems
TCBG Support:
Technical advice, VMD inputs for display.
What will be demonstrated:
A simulation of GlpF (see below) will be shown using
VMD rendered via a
Sun Fire Visual Grid system. The system consists of two V880z computers,
each containing two XVR-4000 graphics boards, that are connected to a
Sun Fire 6800 computer via a high bandwidth multiple Myrinet connection.
The molecular graphics will be displayed on four flat-panel displays in
a 2x2 configuration.
NAMD and VMD are developed by the Theoretical and Computational Biophysics
Group at the University of Illinois. VMD development is supported by
the NIH National Center for Research Resources and the
National Science Foundation.
InfiniCon Systems InfiniBand Opteron Cluster Demo
Title:
NAMD and VMD: Live Molecular Dynamics Simulation on a
InfiniBand Opteron Cluster
Location:
InfiniCon Systems booth
Times:
various, unknown
Investigators:
Klaus Schulten, Robert Skeel, Laxmikant Kale
Developers:
James Phillips, John Stone, Justin Gullingsrud, Gengbin Zheng, Paul Grayson
Presenter:
InfiniCon Systems personnel
Contacts:
www.infinicon.com
TCBG Support:
Technical advice, NAMD and VMD inputs
What will be demonstrated:
A NAMD simulation of GlpF (see below)
will be running on a 16-node, 10Gbps InfiniBand Opteron cluster.
VMD will display the live simulation.
NAMD and VMD are developed by the Theoretical and Computational Biophysics Group at the
University of Illinois. VMD development is supported by
the NIH National Center for Research Resources
and the National Science Foundation.
About the GlpF Demo Simulation (In Both Sun and InfiniCon Booths)
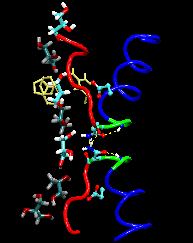
Aquaporins are channel proteins abundantly present in all life forms,
for example, bacteria, plants, and in the kidneys, the eyes, and the
brain of humans. These proteins conduct water and small molecules, but
no ions, across the cell walls. Their defective forms are known to
cause diseases, e.g., diabetes insipidus, or cataracts. The
molecular modeling program, NAMD, along
with large parallel computers at the Pittsburgh and Illinois supercomputing centers
permitted researchers now to model aquaporins in the natural
environment of membrane and water in one of the largest molecular
dynamics simulations ever (over 100,000 atoms). The simulations
revealed in unprecedented detail how cells conduct water and glycerol, a molecule that serves cells' metabolism. The
simulations provided a movie of the entire conduction process.
A greatly reduced model of a single GlpF channel, consisting of only
4210 atoms, 3295 of them fixed, will be used to demonstrate the power
of NAMD to accelerate even small simulations on modest numbers of processors.
The Tcl scripting capabilities of NAMD are used to apply a constant force,
driving a single glycerol molecule back and forth through the channel.
While the force biases the movement of the glycerol, it still takes over
100 ps of dynamics to find the proper alignment to pass through the
specificity filter region of this remarkable molecule.





