NAMD and VMD at SC2001

|
SC2001 Demonstrations
Monday 7pm-9pm
Tuesday 10am-6pm
Wednesday 10am-6pm
Thursday 10am-4pm
by the NIH National Center for Research Resources.
|
Theoretical Biophysics (TBG) and Sun Microsystems Demo
 "If I could just get my hands on that protein!" Single molecule
manipulation techniques like atomic force microscopy have brought us
closer to this frequently expressed wish. These techniques, however,
do not "see" the atomic level detail needed to relate mechanism to
protein architectures. True, computational methods do illuminate the
elusive protein structures, but are limited to static structures, or
trajectories yielded by weeks-long simulations. Now, with
the advent of high-performance computing, interactive
manipulation of molecular dynamics simulations has become a reality.
Linking advanced
molecular graphics
with ongoing
molecular dynamics
simulations, and utilizing a
haptic device
to connect forces from a
user's hand with forces in the simulation, researchers can interact
with "live" proteins. The new methodology is described in a recent
publication
and the figure shown here demonstrates a Cl- ion being
pulled through a gramicidin A channel
(see a 3.8mb Streaming Video).
"If I could just get my hands on that protein!" Single molecule
manipulation techniques like atomic force microscopy have brought us
closer to this frequently expressed wish. These techniques, however,
do not "see" the atomic level detail needed to relate mechanism to
protein architectures. True, computational methods do illuminate the
elusive protein structures, but are limited to static structures, or
trajectories yielded by weeks-long simulations. Now, with
the advent of high-performance computing, interactive
manipulation of molecular dynamics simulations has become a reality.
Linking advanced
molecular graphics
with ongoing
molecular dynamics
simulations, and utilizing a
haptic device
to connect forces from a
user's hand with forces in the simulation, researchers can interact
with "live" proteins. The new methodology is described in a recent
publication
and the figure shown here demonstrates a Cl- ion being
pulled through a gramicidin A channel
(see a 3.8mb Streaming Video).
Theoretical Biophysics (TBG) Talk in PSC Booth
Theoretical Biophysics (TBG) and NCSA Itanium Cluster and Display Wall Demo
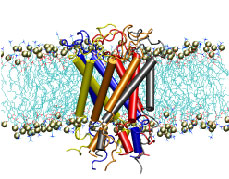 "How do you feel?" Biologists now have an answer that may
surprise you. Our sense of touch relies upon the fact that cells in
our fingertips can sense the pressure from a tabletop and
transmit a signal to the brain. But until recently, the molecular
mechanism for turning the stretching of a cell membrane into a
cellular signal was unknown. An important step in understanding
this process was the discovery of a protein known as a the
mechanosensitive channel of large conductance, or MscL.
Though this protein has been studied primarily in bacteria,
homologues exist in all major kingdoms of life. Researchers in
the Theoretical Biophysics Group have used molecular dynamics
simulations to study, at the atomic level, how MscL opens in
response to pressure changes. Models of MscL will give us new
insight, not only into how we feel, but also how our hearts beat
and how we keep our balance. Feel better now?
(more,
publication)
"How do you feel?" Biologists now have an answer that may
surprise you. Our sense of touch relies upon the fact that cells in
our fingertips can sense the pressure from a tabletop and
transmit a signal to the brain. But until recently, the molecular
mechanism for turning the stretching of a cell membrane into a
cellular signal was unknown. An important step in understanding
this process was the discovery of a protein known as a the
mechanosensitive channel of large conductance, or MscL.
Though this protein has been studied primarily in bacteria,
homologues exist in all major kingdoms of life. Researchers in
the Theoretical Biophysics Group have used molecular dynamics
simulations to study, at the atomic level, how MscL opens in
response to pressure changes. Models of MscL will give us new
insight, not only into how we feel, but also how our hearts beat
and how we keep our balance. Feel better now?
(more,
publication)
This is a simulation of a model of MscL from E. coli provided by our collaborators S. Sukharev and R. Guy from the U. of Maryland and the NIH, respectively. The SMD simulation is designed to test a proposed model of MscL gating.
Theoretical Biophysics (TBG), NCSA and Intel McKinley Demo
 What will be demonstrated:
NAMD will run on two to four McKinley processors. The live simulation
will be displayed with VMD running on a Linux PC.
NAMD and VMD are developed by the Theoretical Biophysics Group at the
University of Illinois. NAMD and VMD development is supported by
the NIH National Center for Research Resources.
What will be demonstrated:
NAMD will run on two to four McKinley processors. The live simulation
will be displayed with VMD running on a Linux PC.
NAMD and VMD are developed by the Theoretical Biophysics Group at the
University of Illinois. NAMD and VMD development is supported by
the NIH National Center for Research Resources.
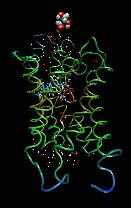
Aquaporins make up an important family of proteins that allow water to pass through cell membranes, while blocking undesired ions and larger molecules. At least eleven different types of aquaporins can be found in the human body, and their malfunction may be linked to common diseases. Some of the aquaporins serve a double-purpose: they also allow the movement of one or more other specific molecules into and out of cells.
This is a simulation of GlpF, a typical aquaporin that also conducts glycerol (a simple sugar), but lets almost nothing else through. A molecule of glycerol is being pulled through the channel, allowing us to see the path that it follows. By studying simulations of this channel, TBG researchers are learning where this channel gets its amazing filtering ability.
BioCoRE Access Grid Presentation



