Lipid Scrambling in nhTMEM16
nhTMEM16 - a dual function lipid scramblase and ion channel
From bacteria to mammals, different phospholipid species are segregated between the inner and outer leaflets of the plasma membrane by ATP-dependent lipid transporters. Disruption of this asymmetry by ATP-independent phospholipid scrambling is important in cellular signaling, but its mechanism remains incompletely understood. Using MD simulations coupled with experimental assays, we show that the surface hydrophilic transmembrane cavity exposed to the lipid bilayer on the fungal scramblase nhTMEM16 serves as the pathway for both lipid translocation and ion conduction across the membrane. Ca2+ binding stimulates its open conformation by altering the structure of transmembrane helices that line the cavity. We have identified key amino acids necessary for phospholipid scrambling and validated the idea that ions permeate TMEM16 Cl- channels via a structurally homologous pathway by showing that mutation of two residues in the pore region of the TMEM16A Ca2+-activated Cl- channel convert it into a robust scramblase.
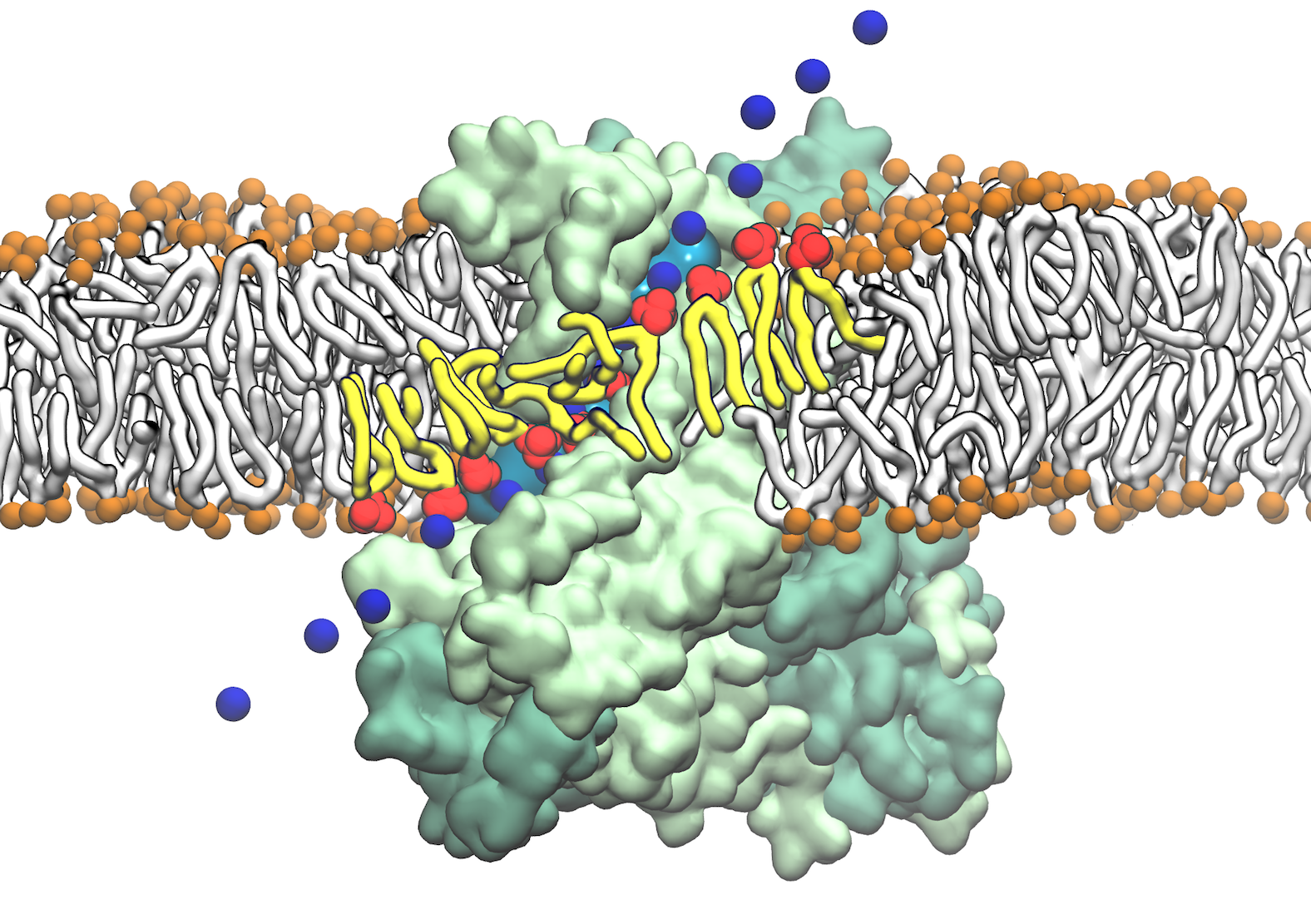
Fig. 1 Direct involvement of phospholipids in biological membrane function, mediated by intimate interactions between lipids and nhTMEM16 scramblase. Lipids lining the hydrophilic aqueduct on the surface of the nhTMEM16 scramblase play a structural role in forming a "proteolipidic" pore, which is likely to be used by ions to cross the membrane. The lipids interacting closely with the protein and coordinating ion permeation are shown in red and tails in yellow. The permeating Na+ ions are shown in time series snapshots (blue spheres). The hydration of the "proteolipidic" pore is illustrated by a transparent cyan surface.
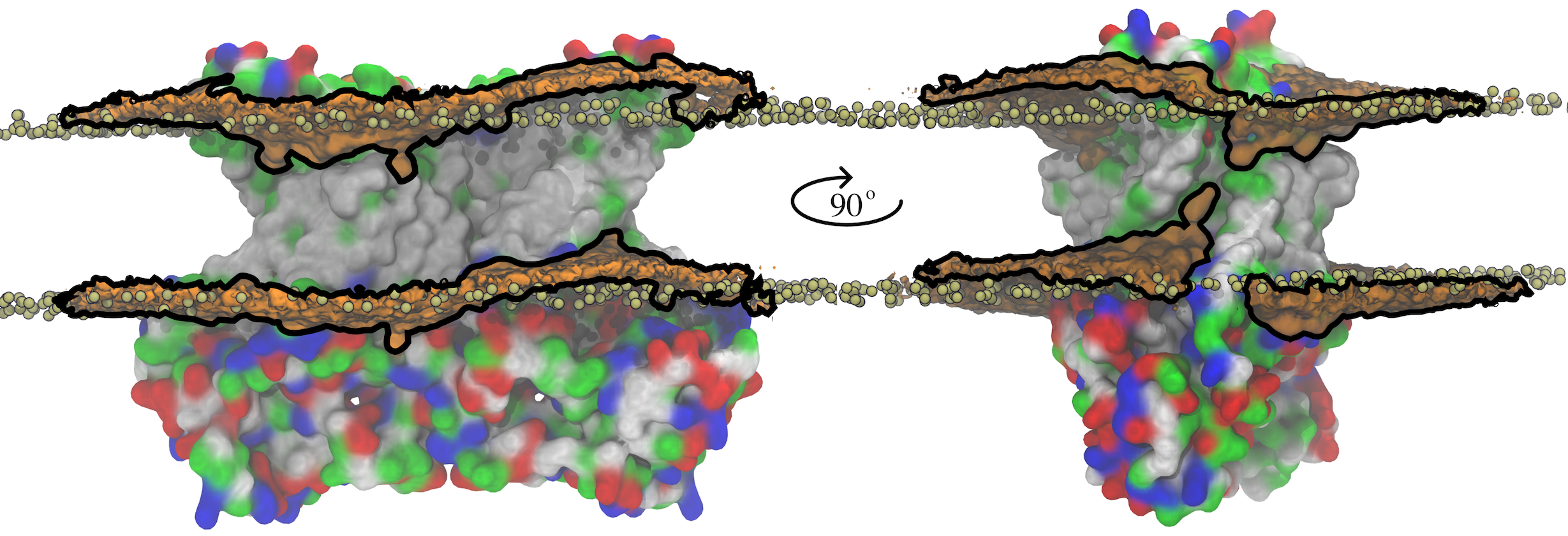
Fig. 2 Membrane deformation induced by nhTMEM16. The initial (t = 0) distribution of phospholipids is shown by the tan spheres representing lipid phosphorus atoms. At the end of the 1000-ns MD simulation, the average phospholipid phosphate density within 10 Å of the protein is shown as an orange surface outlined by black lines that is contoured at 7.5% of the bulk phosphate density.
Membrane deformation induced by the scramblase.
Integral membrane proteins contain hydrophobic segments that are in contact with the acyl chains of the lipid bilayer. From an energetic perspective, the length of the lipid-exposed hydrophobic segments would be expected to approximately match the hydrophobic bilayer thickness. However, in nhTMEM16, the aqueduct that faces the membrane bilayer is strongly hydrophilic, which causes a distortion in the distribution of bilayer phospholipids due to their tendency to match the hydrophobic surface of the protein (Figure 2). This not only reduces the membrane thickness near the aqueduct but also produces a bend in the bilayer centered on the aqueduct. Because the rate of diffusion of a molecule across the membrane is inversely related to the distance (Fick's Law), membrane thinning will prime lipid translocation by reducing the energy required to move a lipid across the membrane. Membrane bending may also favor non-bilayer phases that facilitate scrambling.
Lipid translocation mediated by the scramblase.
Spontaneous penetration of lipid head groups into the membrane interior along the aqueduct was observed during the simulation, resulting in a continuous file of lipids connecting the outer and inner leaflets (Figure 3). Full permeation of a lipid from the inner leaflet to the outer leaflet of the membrane was captured during the equilibrium simulation (Movie 1 and 2). Four more full translocations of lipids (including charged lipid POPS) took place when transmembrane voltage was applied.
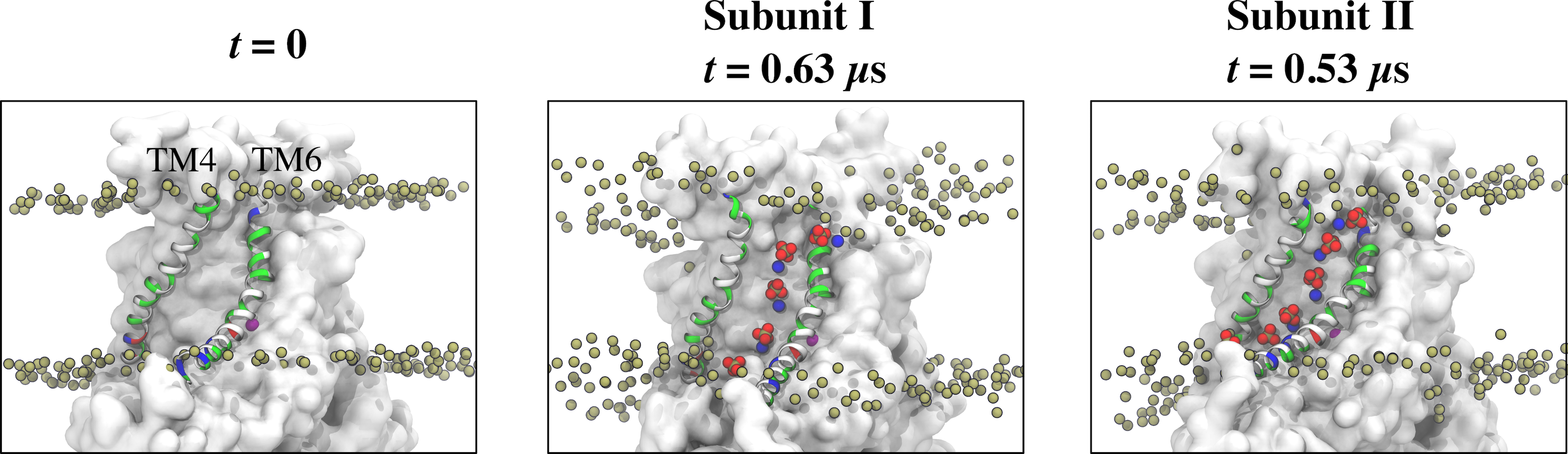
Fig. 3 Representative snapshots of the aqueduct during an MD simulation with Ca2+-activated nhTMEM16. The aqueduct is devoid of phospholipid head groups at t = 0 (left panel), but becomes occupied by lipids in both subunits during the simulation.
Movie 1. Full scrambling of a POPC lipid from inner leaflet to outer leaflet of the membrane through the hydrophilic aqueduct on the surface of nhTMEM16 scramblase under equilibrium condition. The simulation is 950 ns in duration.
Ion permeation occurs through the same structural pathway of lipid scrambling.
In the simulations, ion conduction through the aqueous pore formed between the protein and the lipid head groups developed in parallel with phospholipid transport (Figure 4 and movie 2) and became more robust as lipids became more tightly packed and organized in the aqueduct as scrambling proceeded. In this flexible pore structure, lipids play a structural role by lining the hydrophilic ion conduction pathway with their head groups. The simulations also show that the conduction pathway is at least partly hydrated and that permeant ions are coordinated by water molecules a majority of their transit time across the membrane.
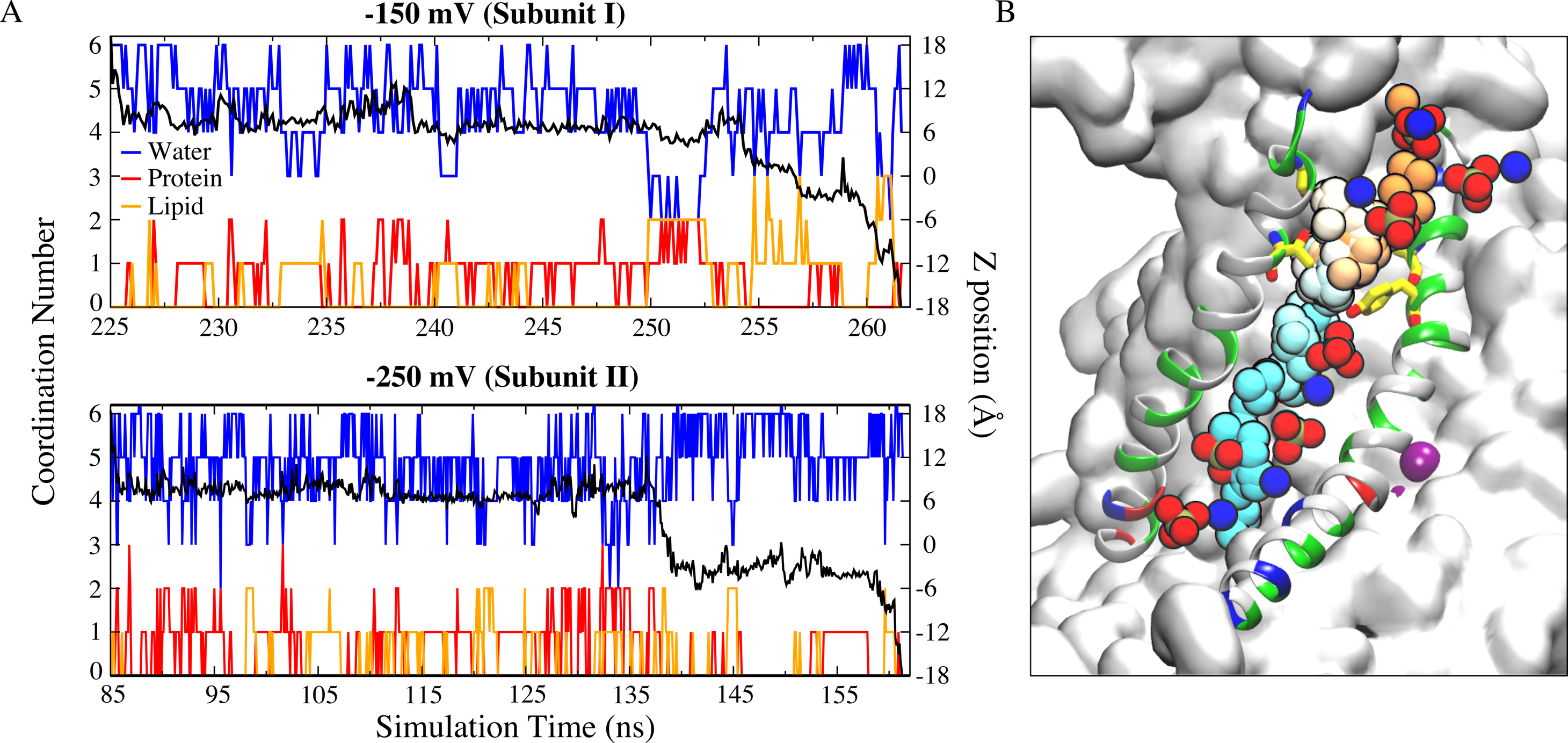
Fig. 4 (A) Full permeation of Na+ ion from the extracellular side to the intracellular side of the membrane was captured during the simulations at −150 mV (top panel) and −250 mV (bottom panel). Oxygen atoms from protein (red) and lipid head groups (orange) both contribute to the ion coordination throughout their permeation. The black curve is the corresponding z-position of the permeant ion. (B) Time series snapshots representing translocation of the Na+ ion with time represented in pseudo-color from orange (start) to cyan (end). The lipid head groups lining the ion permeation pathway are shown as phosphate groups and nitrogen atoms.
Movie 2. Ion permeation through the "proteolipidic" pore was captured during a simulation on nhTMEM16 scramblase under transmembrane potential. Lipids play a structural role by lining the hydrophilic ion conduction pathway with their headgroups.
Related Publication
-
Lipids and ions traverse the membrane by the same physical pathway in the nhTMEM16 scramblase. Tao Jiang, Kuai Yu, H Criss Hartzell, and Emad Tajkhorshid. eLife, 2017. DOI: 10.7554/eLife.28671
Investigators
Page created and maintained by Tao Jiang.



