DNA with analogue bases
DNA hydration has been studied extensively, both experimentally and theoretically. Computational studies on the hydration of DNA showed conformation-dependent differences in both geometry and the extent of hydration for the major and minor grooves. The role of water-mediated hydrogen bonds in protein-DNA interactions and the potential contribution to the thermodynamics of DNA-ligand binding indicates the need to know the precise location of water molecules bound in the minor and major grooves of DNA.
Nucleoside analogues offer an alternative means to explore the role of water molecules in DNA and in protein--DNA interactions. J. Rockhill [1] suggested that the analogue 2'-deoxy-7-hydroxymethyl-7-deazaadenosine (hm7c7dA), might mimic a bound water molecule complexed to an adenine residue in the major groove of DNA.
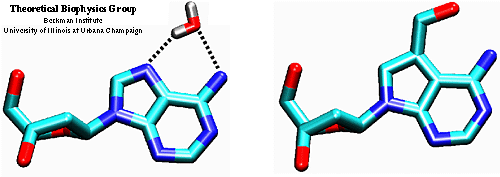
Adenosine (left) and 2'-deoxy-7-hydroxymethyl-7-deazaadenosine (hm7c7dA) (right). Coloring scheme for atoms: oxygen is red, carbon is cyan, nitrogen is blue.
The analogue (hm7c7dA) was modeled into the DNA dodecamer, substituting the two adenine bases in one strand (A5 and A6) individually or jointly, creating four distinct systems.
The simulations, when compared to those of the dodecamer itself, show that incorporation of the analogue affects neither the overall DNA structure nor its hydrogen bonding and stacking nteractions when a single individual base is replaced by the analogue.
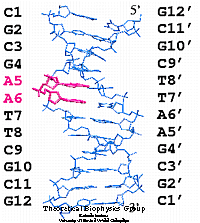
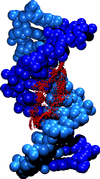
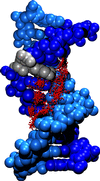
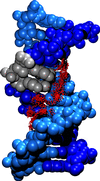
Crystal structure of the DNA dodecamer used in the simulation and numbering scheme for the DNA bases. The adenine bases substituted individualy or jointly with the analogue are represented in red.
Water molecules near the bases in the singly-substituted oligonucleotides are similarly unaffected. Double substitutions lead to differences in all the aforementioned parameters with respect to the reference sequence.
Water molecules within hydrogen bonding distance (proton-acceptor distances < 2.5 A and angle donor-proton-acceptor > 120 to the acceptor/donor groups of the GAATTC bases were selected and are shown superimposed (for a better representation the oxygen atoms of the water molecules are shown as vdW sphere with a reduced radius). The DNA atoms are in vdW representation. The analogue bases are shown in grey.(click on the pictures for bigger images)
The results suggest that the analogue provides a good mimic of specific "ordered" water molecules observed in contact with the DNA itself and in protein-DNA complexes.