Unfolding fibronectin type III modules
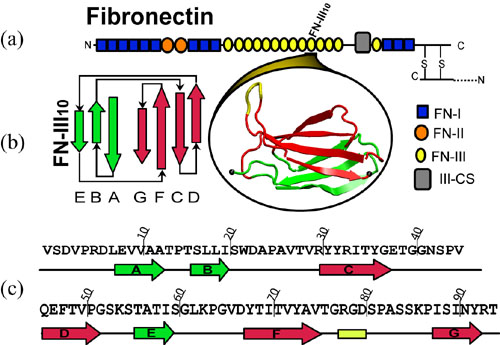
Figure 1
Fibronectin (FN) molecules are one example of mechanical proteins because they are exposed to mechanical tension in the extracellular matrix environment. Transmitted through transmemberane receptor intergrins, mechanical forces were used to assemble fibronectins into fibrillar networks, which couple cells mechanically to their environment and to neighbor cells. Structurally, FN is a 450 KDa dimer composed of homologous, repeating structural motifs (classified as fibronectin repeats FN-I, FN-II, and FN-III) that are grouped together into functional domains. Figure 1 shows the diagram of a FN monomer, and the structure and seqence of the 10th type III FN module, FN-III-10. Similar to muscle protein titin I27, the FN-III modules possesses a beta-sandwich structure consisting of seven beta-strands (A-G) that are arranged in two anti-parallel sheets ABE and FGCD. FN-III-10 is of special interests because it mediates cell adhesion to surfaces via its integrin binding motif, Arg, Gly, and Asp (RGD), that is located at the apex of the loop shown in yellow in Fig. 1(b).
In collaborating with the research group of Viola Vogel of the Department of Bioengineering at the U. of Washington in Seattle, we have used SMD to simulate the unfolding processes of FN-III modules. Fig. 2 shows a FN-III-10 module solvated and stretched in a periodic water box, consisting of 126,082 atoms in total.
Figure 2
Three different unfolding pathways of FN-III-10 has been identified in SMD simulations, as shown in Fig. 3. Discussed in an earlier publication, the separation of G-strand from the remaining part resulted in a gradual shortening of the distance between the apex of the RGD-containing loop and the module surface, which reduces the loop's affinity to surface-bound integrins. The RGD-loop is therefore strategically placed to undergo strong conformational changes and constitutes a mechanosensitive switch for recognition by integrin receptors. The separation of G-strand can happen at very short extension of about 20 A (mpeg animation, 802K) or at longer extension of about 100 A, depending on which pathway the protein selects. Fig. 4 shows a typical extension force profile from a constant force SMD stretching of FN-III-10.
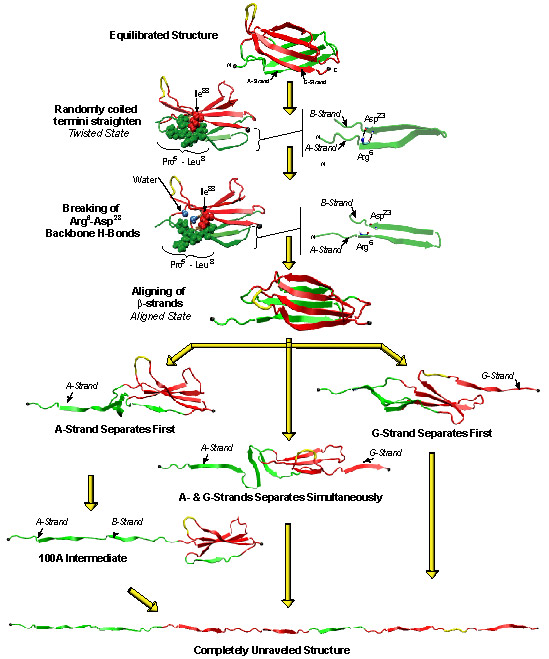
Figure 3
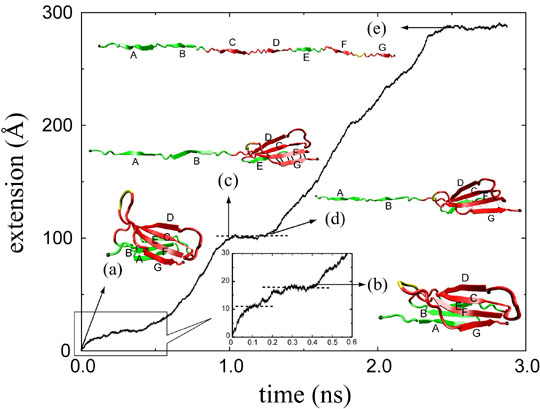
Figure 4
Systematic constant force SMD simulations have been conducted with fibronectin monomers FN-III-7 to FN-III-10. The mechanical stability of FN-III-10 is the weakest among these four modules. Two distinct kinetic barriers, seen as plateaus in extension-time profile from constant force SMD simulations, have been identified for short stretches less than 60 A: the first barrier corresponds to straightening of a conserved beta-bulge and the disruption of the hydrophobic packing of the two beta-sheets, when the protein moves from a twisted orientation to an aligned orientation of the two beta-sheets (see Fig 4(b)); the second barrier corresponds to the rupture of a patch of interstrand hydrogen bonds between two beta-strands.
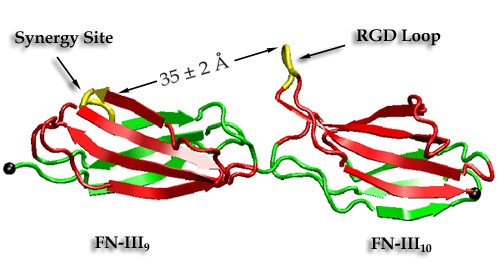
Figure 5
FN-III-9 contains a synergy site which enhance the intergrin binding of RGD loop on FN-III-10. A recent report described how mechanical stretching affects the relative distance between these two synergy-RGD sites (see Fig. 5). Prior to unfolding of the modules, the simulations revealed an intermediate state in which the synergy-RGD distance is increased to approximately 55 A while the conformations of both sites remained unperturbed. Since it has also been experimentally found that increasing the linker chain between FN-III-9 and FN-III-10 reduces integrin binding, this distance is too large for both sites to co-bind the same receptor. Our simulations thus suggest that increased integrin-binding contributed by the synergy site and associated downstream cell-signaling events can be turned off mechanically by stretching FN-III-9-10 dimer into this intermediate state.



